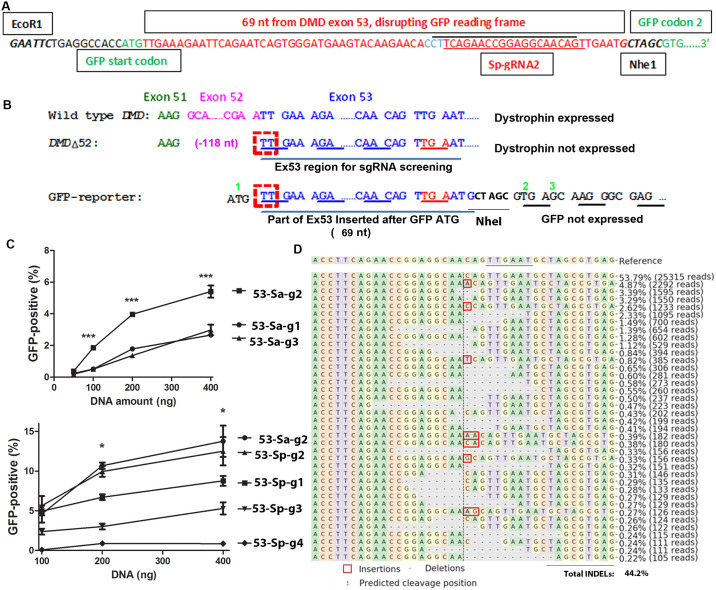
Fig 2. A GFP-reporter assay for evaluating sgRNAs for DMD reframing by single sgRNA cut after exon 52 deletion.
A. Design of the GFP-reporter cassette. The first and the second codons of GFP are shown green. The inserted sequences from DMD exon 53 are shown in red. The target sequences for Sa-gRNA2 (below the black line) and for Sp-gRNA2 (above the red line, the PAM region is shown in blue) are indicated. The two restriction enzyme sites added for sub-cloning are also shown. B. Comparison of the reading frames of the disrupted GFP and DMD. The normal dystrophin reading frame (top) and the reading frame shift caused by exon 52 (pink) deletion are shown. The insertion of 74 nt (69 nt from exon 53 and the rest 5 nt to make a NheI enzyme site for possible other target cloning) after the GFP start codon creates the same frame shift to GFP as the deletion of exon 52 to dystrophin. A deletion of 3n+2 nucleotides (exampled by the red dashed boxes when n = 0) will restore reading frame for both. Green numbers indicate amino acid numbers for the wild type GFP. C. Comparing SaCas9 (top) and SpCas9 gRNAs (bottom) by transfecting GFP-reporter cells. *** indicates p<0.0001 when Sa-gRNA2 was compared with other Sa-gRNAa (ANOVA). * indicates p<0.05 when Sa-gRNA2 was compared with SpCas9 gRNAs other than Sp-gRNA2 (ANOVA). Increasing amount of DNA was transfected into 1.25x105 GFP-reporter cells and the cells were analyzed by flow cytometry 48 hours after transfection. Three replicates were included for each condition. Shown were representative data of at least two independent experiments. D. Next generation sequencing (NGS) analysis of INDELs in the target region of GFP-reporter cassette. The GFP-reporter cells (1.25x105) were transfected with 0.5 μg plasmid DNA co-expressing SaCas9 and sgRNA Sa-gRNA2. The target DNA in the GFP-reporter cassette was amplified with primers (reporter-F1 and reporter-R2) for sequencing analysis. The original sequence (reference) is listed on the top line with the protospacer adjacent motif (PAM, NNGRRT for SaCas9, “N” can be any nucleotide, and “R” is A or G) underlined. Below the reference sequence are listed types of readings over 0.2% observed in NGS. Reading number and percentage of each type of reading are listed at the right of that sequence. Note that all INDELs are around the predicted cleavage site (vertical dashed line).