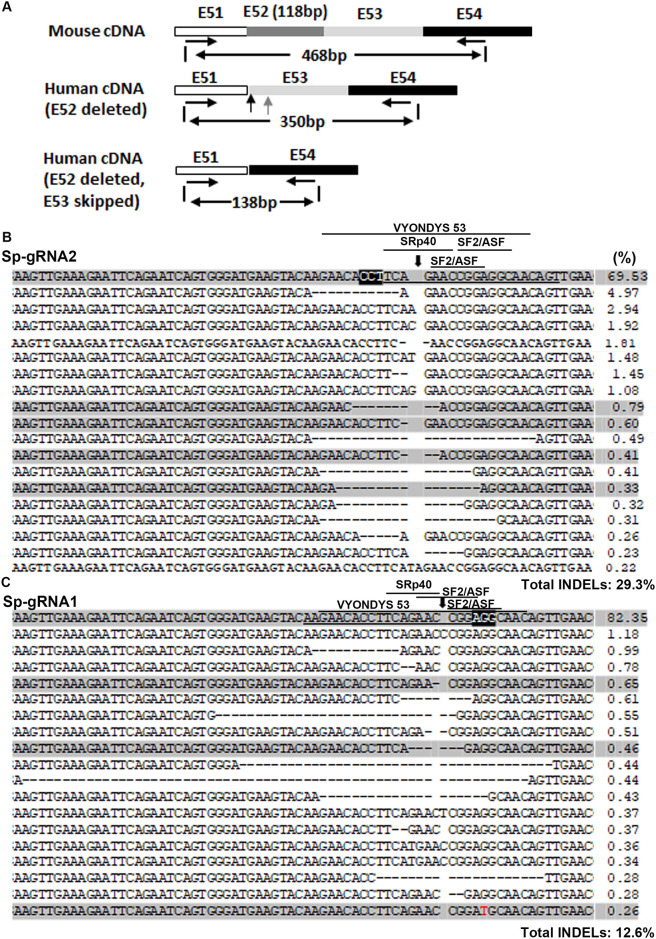
Fig 4. NGS analysis of cDNA from hDMDdel52/mdx myoblasts treated with various RNPs.
A. Diagram showing the strategy of PCR amplification of human DMD cDNA for NGS. The forward and reverse PCR primers were DMD51-EX-F and DMD54-R1. PCR products of 100–350 bp were recovered for NGS, so that the mouse Dmd derived PCR products (468 bp) were not analyzed. B. Analysis of cDNA sequences without exon 53 skipping after Sp-gRNA1 RNP treatment. C. Analysis of cDNA sequences without exon 53 skipping after Sp-gRNA2 RNP treatment. For (B and C), myoblasts (1x105) were treated with each 120 ng p24 of LVLP-RNPs. The human DMD target sequence was amplified by strategy shown in A for NGS. The PAM regions are highlighted black and the target sequences are underlined. The predicted cleavage sites are indicated by a vertical arrow. Dashed lines indicate deletions. Substitutions are in red. Numbers at the right are the percentages of the alleles. The shaded sequences are those did not reframe dystrophin. The predicted high score ESE motifs and the target region for VYONDYS 53 (golodirsen) were indicated by lines.