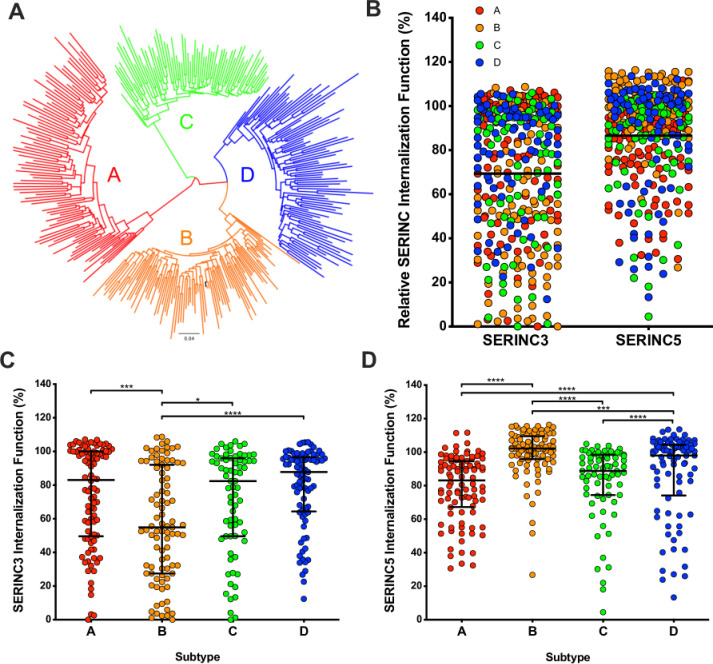
Fig 1. Nef-mediated SERINC3 and SERINC5 internalization function varies among circulating Nef alleles representing globally relevant subtypes.
(A) A maximum likelihood phylogenetic tree of 339 participant-derived HIV Nef clones is shown. Subtype A clones are represented in red, subtype B in orange, subtype C in green and subtype D in blue. The subtype B NL4.3 Nef strain (black) was included as a reference. (B) The relative ability of all Nef clones to internalize SERINC3 (left) or SERINC5 (right) is shown. SERINC internalization was assessed by flow cytometry following co-transfection of CEM T-cells with Nef and SERINC(iHA) (also see S1 Fig), and values were normalized to that of the NL4.3 Nef control (set as 100%). (C) The relative ability of each Nef clone to internalize SERINC3 is shown, stratified by subtype. (D) The relative ability of each Nef clone to internalize SERINC5 is shown, stratified by subtype. Bars represent the median and the interquartile ranges. Statistical analysis was performed using the Mann-Whitney U-test and significant results are indicated by asterisks (* = p<0.05; ** = p≤0.01; *** = p≤0.001; **** = ≤0.0001).