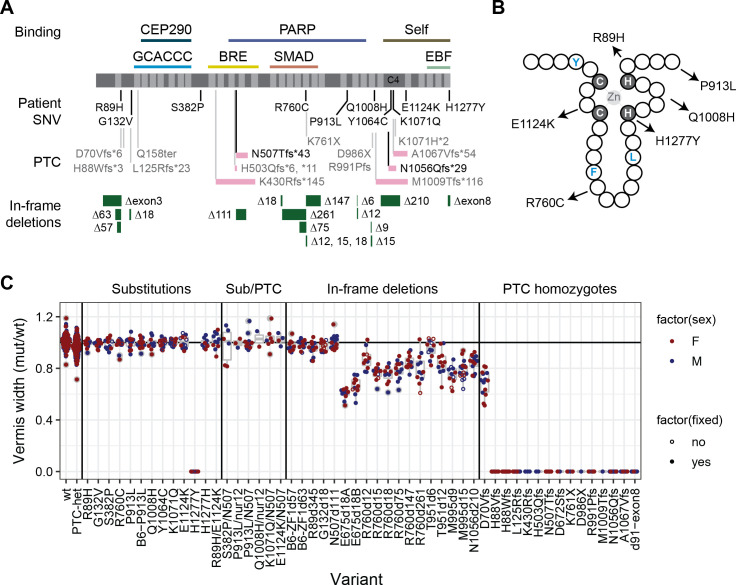
Fig 1. Zfp423 mutations induced by genome editing affect vermis size.
(A) Locations of specific variants are indicated relative to ZNF423 RefSeq protein NP_055884 (dark grey), C2H2 zinc fingers (light grey), and reported binding activities. Frame-shift PTC variants show relative length of altered reading frame (pink bars). N507Tfs*43 and N1056Qfs*29 were humanized to encode the same frame-shifted peptide as reported patient variants. For in-frame deletions (green bars), the number of deleted base pairs is shown. (B) Schematic of a C2H2 zinc finger shows relative positions of patient substitution alleles relative to consensus hydrophobic (blue) residues and required zinc-coordinating residues (grey background). (C) Ratio of mutant to control vermis width measured from surface views of same-sex littermate pairs. Each dot represents one littermate pair. Female pairs, red, male pairs, blue. A few pairs were measured as freshly dissected, unfixed material (open circles), which did not noticeably affect the relative measure. PTC heterozygous, H1277Y homozygotes, homozygotes for each in-frame deletion except N507d111, and all PTC homozygous variants were significantly different from both the expected null model and empirical wild-type:wild-type comparisons.