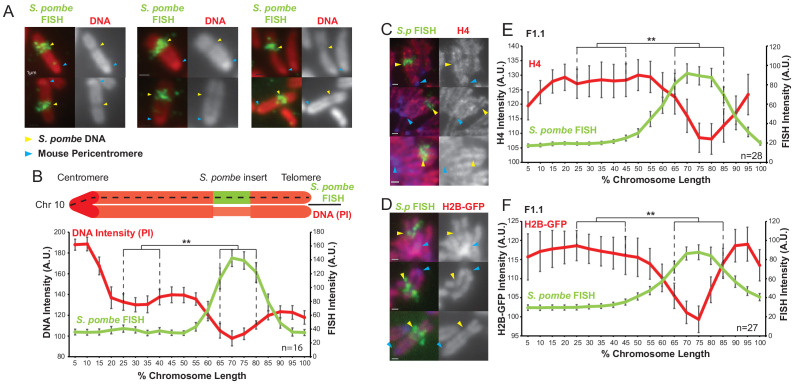
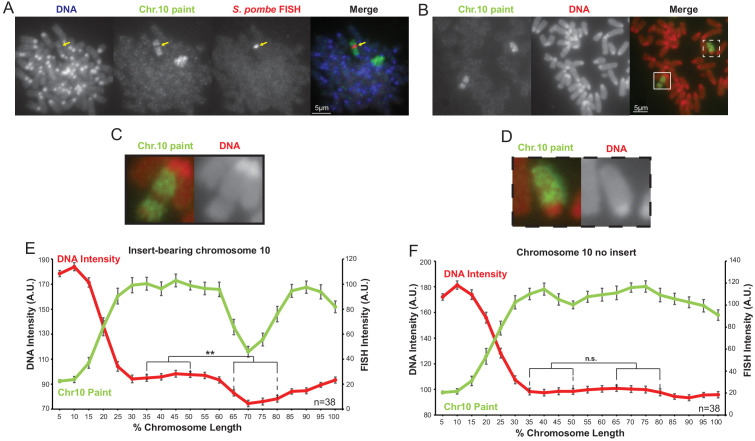
Figure 1. S. pombe DNA inserted into a mouse chromosome adopts a distinct structure on mitotic chromosomes.
(A) Metaphase spreads of mouse F1.1 chromosomes showing the distinct structure of the S. pombe DNA insert. Propidium iodide stained DNA (PI, red), S. pombe DNA FISH using probes from total S. pombe DNA (green – yellow arrows), centromeres (regions of brighter DNA staining - blue arrows). Scale bars: 1 μm. (B) Schematic representation and average chromosome profile of the F1.1 insert-bearing chromosome across several images (n = 16, Figure 1—source data 1). Signal intensities of PI DNA stain (red) and FISH signal (green) were measured along the length of the chromosomes and binned according to their position, from the centromere (0–5%) to the telomere (95–100%). Error bars represent ± standard error from the mean (SEM). Average DNA stain intensity was compared between the regions of 25–40% (endogenous mouse DNA) and 65–80% (S. pombe DNA corresponding to the highest FISH signal) by the KS test (**p<0.001). (C and D) Metaphase spreads of F1.1 cells either stained by immunofluorescence for histone H4 (C) or expressing tagged histone H2B-GFP (D) (red), with S. pombe DNA FISH (green) and DAPI-stained DNA (blue). S. pombe DNA (yellow arrows) and centromere (blue arrows) locations are indicated. Scale bars: 1 μm. (E and F) Average signal intensity profile of the F1.1 insert-bearing chromosome showing FISH and either anti-H4 (E, Figure 1—source data 2) or H2B-GFP (F, Figure 1—source data 3) across several images (n = 28, 27). Error bars represent ± SEM. Average histone levels were compared between the regions of endogenous mouse DNA and S. pombe DNA highlighted by FISH by the KS test (**p<0.001).