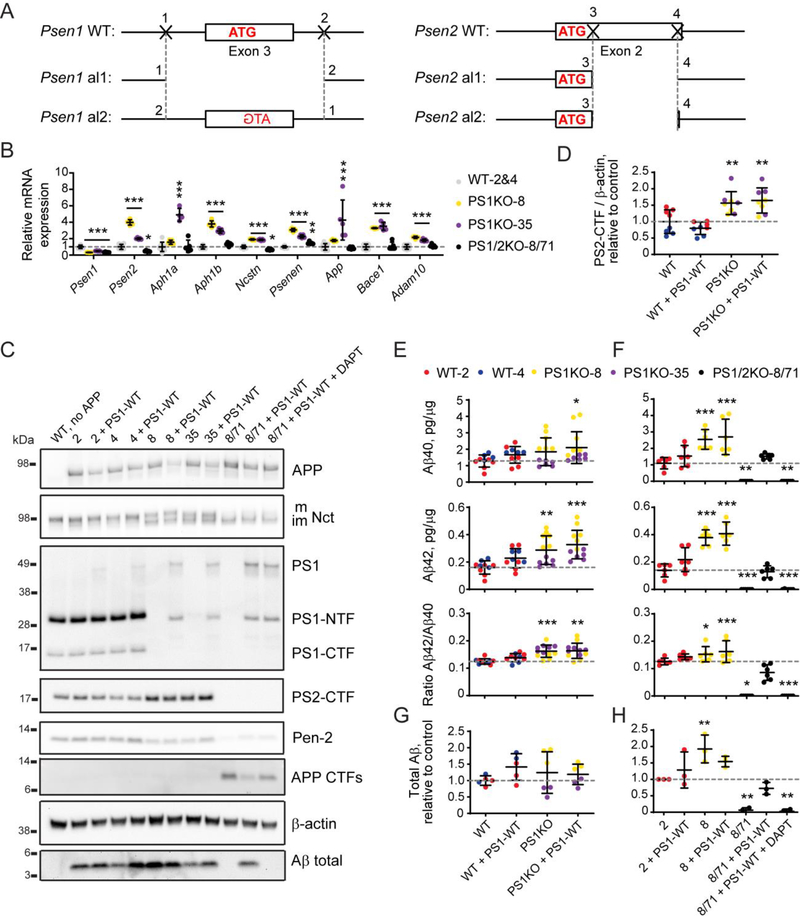
Figure 3. Validation of double Psen1 and Psen2 knock-out in N2A cells and rescue with exogenous PSEN1 expression.
(A) Schematic representation of editing in the genomic loci of Psen1 and Psen2 that created a double knock-out of mouse presenilin 1 and 2 in N2A cells by CRISPR/Cas9-mediated genome engineering. Nucleotides in bold belong to the coding exon. Start codon nucleotides are labeled in red. (B) qPCR for genes of γ-secretase subunits, Adam10, Bace1, and App in wild type (2 and 4) and knock-out (8, 35 and 8/71) clones. (C) Analysis of γ-secretase subunits and APP processing by western blotting in wild type (2 and 4) and knock-out (8, 35 and 8/71) clones rescued with wild type human presenilin 1 (PS1-WT). Quantification of western blotting data is in Supplementary Figure S1A. (D) Quantification of endogenous presenilin 2 protein levels in N2A-PS1KO clones (8 and 35) versus wild type N2A clones (2 and 4) based on (C). (E - H) Quantification of Aβ40, Aβ42, Aβ42/Aβ40 ratio by ELISA (E, F), and total Aβ by western blotting (G, H) in PS1KO clones (8 and 35) versus wild type N2A clones (2 and 4) (E, G) and in PS1/2KO-8/71 cells versus the wild type (2) and parental (8) lines (F, H). Values shown are mean ± SD of n = 3 independent experiments with one (D, G, H) or two (E, F) technical replicates each. * P < 0.05, ** P < 0.01, *** P < 0.001, one-way ANOVA with Dunnett’s post-hoc test against the control condition (WT in D, E, G and 2 in F, H).