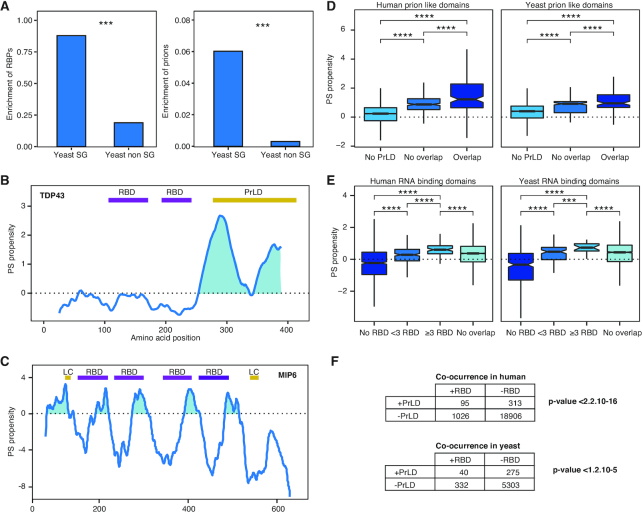
Figure 1.
RBDs and prions are associated with the proteins PS ability. (A) Stress granules are enriched in RNA-binding proteins (RBPs) and prions. Left, percentage of RBPs (90) found in the yeast SGs (15) and in the rest of the proteome (40). Right, percentage of prion proteins (30) in same datasets. ***: P-value < 0.001; Fisher's exact test. (B) catGRANULE PS propensity profile predicted for human Tdp43. We show two RBDs (Pfam annotated) in purple and a PrLD (PLAAC algorithm prediction) in yellow (42). (C) PS propensity profile of yeast Mip6. We report the four RBDs in purple and two low-complexity (LC) regions in yellow, as annotated at the ELM database (96). The two RBDs close to the N-terminus are essential for the PS (41). (D) Box plots showing the percentage of overlapping between PrLDs and the highest peak of PS predicted by catGRANULE. Overlap: ≥20% PrLD propensity predicted using PLAAC [PLAAC peak (42)]; No overlap: <20%; No PrLD: PLAAC does not predict PrLD. To guarantee a minimum overlap of one amino acid, we chose a threshold of 20% overlapping between peaks, it is important to note that we obtained similar results regardless of the threshold chosen. (E) Box plots showing the percentage of overlapping between RBDs (Pfam annotated, scanned with HMMER) and the highest peak of PS predicted by catGRANULE. Proteins were classified according to the number of RBDs contained in their sequence: >20% overlap with at 1 RBD in ‘<3 RBD’, and ‘≥3 RBD’; those with no Pfam annotation as No RBDS; finally ‘No overlap’ indicates <20% overlap between the RBD and the highest peak of PS. For all box plots: box represents interquartile range (IQR); central line, median; notches, 95% confidence interval; whiskers, 1.5 times the IQR. ****: P-value < 0.0001; t-test. (F) Fisher test of the association between PrLDs and RBDs. Proteome datasets interrogated for presence (+) or absence (−) of PrLDs (predicted with PLAAC) and RBDs (scanned with HMMER, based on Pfam).