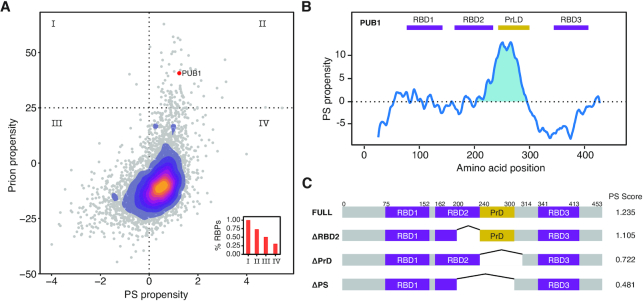
Figure 2.
Pub1 as a model to study PS. (A) Scatter plot and over-imposed 2D-density plot indicating the PS propensity (catGRANULE, X-axis) (19,41) and prion propensity (PLAAC, Y-axis) (42) for the yeast proteome. The confidence thresholds of the predictors are highlighted with dotted lines. Inside, bar plot indicating the percentage of RBPs in each of the quadrants. Pub1 protein (three RBDs) is indicated with a red dot. (B) catGRANULE prediction of Pub1 PS propensity (Y-axis) along sequence (X-axis). The figure shows three RRMs (Pfam annotated, RRM, PF00076 (97)) in purple and a PrLD (PLAAC prediction) in yellow. (C) Diagram of the different Pub1 variants studied in this thesis. Targeted deletions of Pub1 full: ΔPrLD (Δ240–314), ΔRRM2 (Δ200–240) and ΔPS (Δ200–300).