Figure 4.
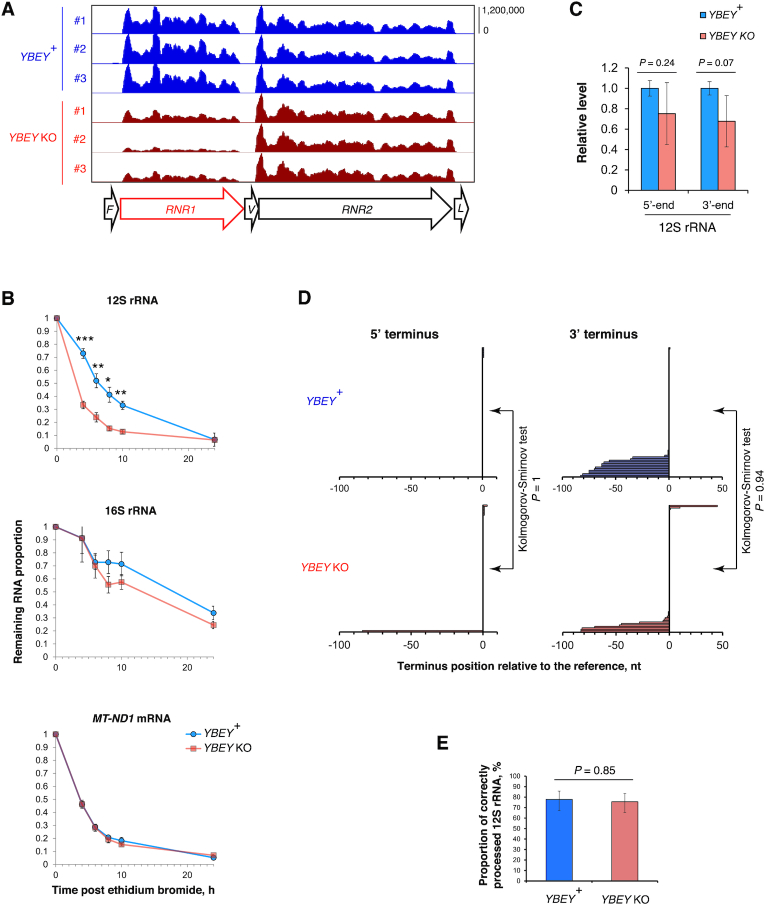
12S rRNA is destabilized, yet correctly processed in the absence of YBEY. (A) Snapshot of the mitochondrial rRNA locus showing a select downregulation of 12S rRNA in the absence of YBEY, as assessed by RNA-seq. tRNA genes are labelled with the single-letter code. (B) The half-life of 12S rRNA is significantly decreased in YBEY KO cells. RNA samples were collected at various time points after arresting mitochondrial transcription with ethidium bromide and analysed by RT-qPCR. Means ± SEM for n = 4 (YBEY+) or n = 9 (YBEY KO) are shown; P-values, two-tailed Welch's test: *P = 0.013, **P < 0.005, ***P = 7.7 × 10−5. (C) RT-qPCR analysis of the 12S rRNA precursor in YBEY+ and YBEY KO cells. The 5′ extension of the primary transcript into tRNAPhe and the 3′ extension into tRNAVal were probed by qPCR across the respective junction. Means ± SEM for n = 3 are shown; P-values, two-tailed t-test. (D) cRT-PCR analysis of the 5′- and 3′-termini of 12S rRNA in YBEY+ and YBEY KO cells. n = 77 plasmid clones from two independent YBEY+ cell lines and n = 86 clones from two independent YBEY KO cell lines, represented by horizontal bars, are aligned with respect to the reference positions (a negative number means that the terminus is upstream, a positive – downstream, and ‘0’ – at the expected reference position). The majority of clones showed correct processing at both ends. (E) YBEY+ and YBEY KO cells have indistinguishable proportions of correctly processed 12S rRNA molecules. Proportions and 95% CIs are shown; based on the data in (D); P-value, Fisher's exact test.