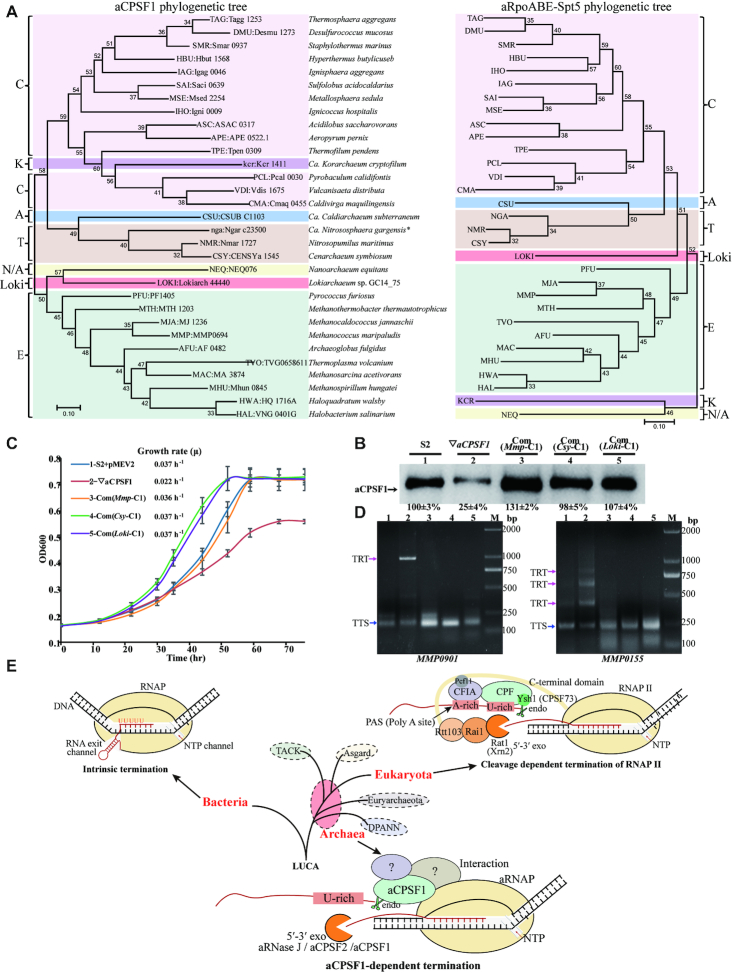
Figure 6.
The ubiquitous distribution of the aCPSF1 orthologs among Archaea and the aCPSF1 dependent archaeal transcription termination reveals a similar 3′-end cleavage mode as the eukaryotic RNAP II termination. (A) The Maximum Likelihood phylogenetic trees based on aCPSF1 proteins (left) and concatenated RNA polymerase subunits A, B and E, and transcription elongation factor Spt5 (right). Protein sequences were retrieved from genomes or metagenome-assembled genomes of the representative archaeal lineages that have been described thus far, including Euryarchaeota (E), Crenarchaeota (C), Thaumarchaeota (T), Aigarchaeota (A), Korarchaeota (K), Nanoarchaeota (N), and Lokiarchaeota (Loki). The unrooted tree was constructed using Maximum-Likelihood methods in MEGA 7.0 software package. Bootstrap values are shown at each node. (B) Protein abundances of the aCPSF1 orthologs in complemented ▽aCPSF1 strains were examined by western blot using the Mmp-aCPSF1 polyclonal antiserum. The percentiles of protein content referenced to that in S2 are shown beneath the gel. (C) Ectopic expression of the aCPSF1 orthologs from Lokiarchaeum GC14_75 (com(Loki-C1)) and Cenarchaeum symbiosum (com(Csy-C1)) restored the 22°C-growth defect of ▽aCPSF1 similarly as the complementation of itself (com(Mmp-C1)) and wild-type S2 carrying an empty vector (S2-pMEV2). The averages of three replicates and standard deviations are shown. (D) 3′RACE assays the TRT transcripts of MMP0901 and MMP0155 in the strains numbered as in panel (B). (E) A proposed mechanism of aCPSF1 cleavage triggering the archaeal transcription termination exposes a homolog of the eukaryotic RNAP II termination mode (3–5). The bacterial intrinsic termination is included for comparison. LUCA means last universal common ancestor.