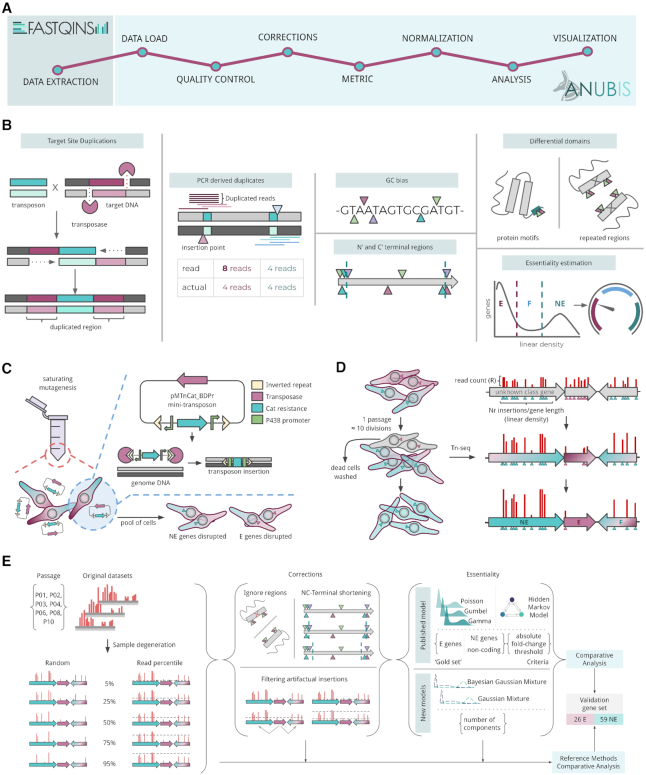
Figure 1.
Graphical abstract. (A) Proposed workflow using FASTQINS to process raw sequencing files into insertion profiles and ANUBIS to explore essentiality-related problems and provide estimates. (B) Graphical representation of the different issues that are not considered in previous essentiality studies. Target site duplications can double the signal of a transposition event (the transposon in blue is flanked by two different chromosome positions that are at a fixed distance equal to the duplication size). Reads derived from the PCR process can artifactually increase the signal of an insertion point (symbolized as triangles). GC content biases can occur when a transposase shows preference for TA sites. At the level of the protein, 5% of the N’- and C’-termini are arbitrarily not considered because they tend to accept insertions with no impact on essentiality. The differential essentiality of protein motifs and a lack of mapping due to repeated motifs should also be considered. Finally, essentiality can be estimated by different models and assumptions. (C) Saturating mutagenesis of M. pneumoniae with the mini-transposon pMTnCat_BDPr, which includes a Tn4001-derived transposase and a Cat resistance marker flanked by P438 promoters. With this approach, E and NE genes are expected to be disrupted in a random manner. (D) The library was selected along 10 serial selection passages (10 cell divisions each). (E) Information was collected from seven different passages (n = 2) and degenerated by two types of sampling. These samples were used to iterate and evaluate different combinations of corrections, essentiality models and criteria. Results were assessed by comparing the level of agreement between estimates and a validated set of 84 genes of known categories.