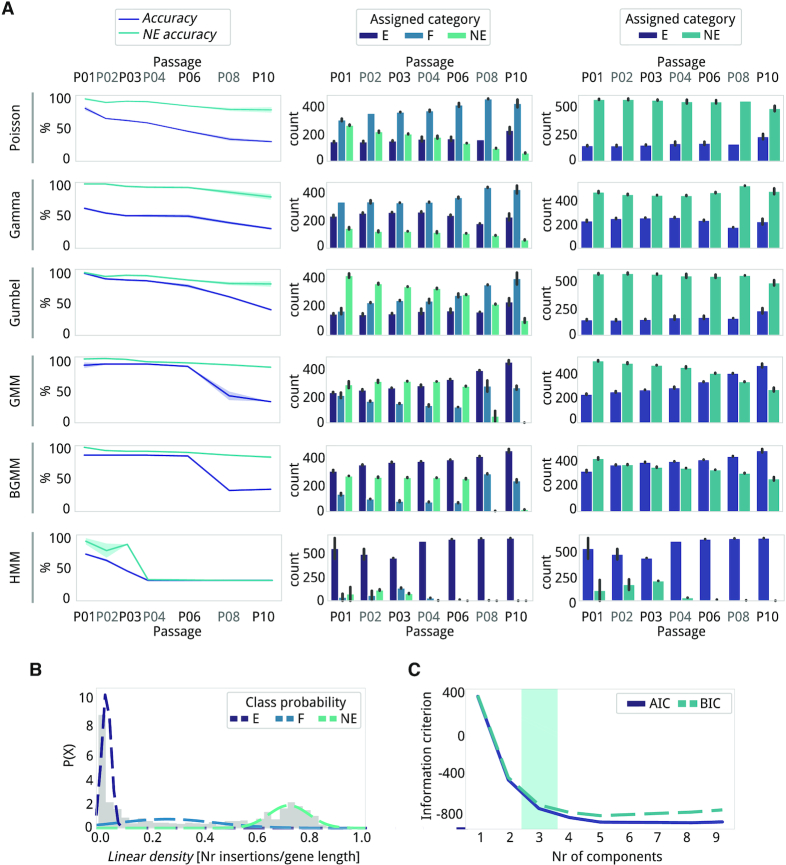
Figure 3.
Comparison of accuracy and gene category assignment between reference and new essentiality estimate models. The methods used are labeled on the left (GMM, Gaussian Mixture Model; BGMM, Bayesian Gaussian Mixture Model; and HMM, Hidden Markov Model). (A) left panel, Accuracy (purple) and NE accuracy (light blue) in percentage values for each method per passage. center panel, Number of genes classified as E (purple), F (blue), and NE (light blue). Error bars represent the standard deviation (n = 2). right panel, Number of genes classified E (purple) and NE (blue), with F and NE genes grouped together. Error bars represent the standard deviation (n = 2). (B) An example of an essentiality estimate using the Gaussian Mixture Model (GMM) with three components for P01, replica 1 (replica 2 in Supplementary Figure S4). The gene linear density (grey histogram) has been properly fitted to the data using three Gaussian distributions (dashed lines: E (purple), F (blue) and NE (light blue)). (C) Akaike Information Criterion (AIC) and Bayesian Information Criterion (BIC). The lower the AIC and BIC values, the better the balance between goodness-of-fit and model simplicity. The number of components (i.e. 3, blue shadowing) represents the elbow of the line where there is a good trade-off between fitting and the number of parameters.