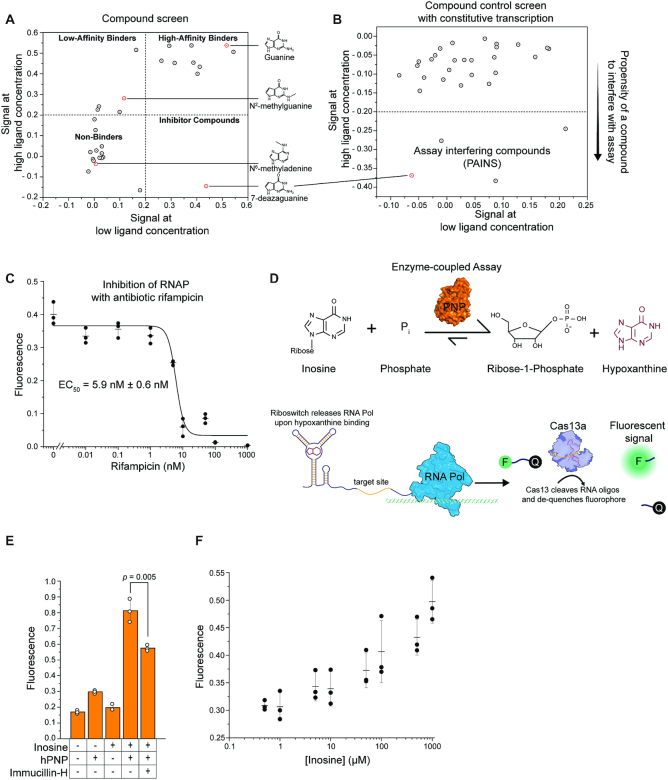
Figure 5.
SPRINT can be used for compound screens and enzyme-coupled assays. Fluorescent signal of the SPRINT reactions was background subtracted and normalized to 125 nM fluorescein. (A) The guanine riboswitch xpt/pbuE*6U regulated transcription in response to 30 different compounds. Signal with solvent only was subtracted from signal with ligand to correct for differences in solvents of ligands. Dots represent mean value, n = 3. (B) The effect of the compounds on transcription from a constitutive promoter was measured. Signal with solvent only was subtracted from signal with ligand to correct for differences in solvents of ligands. Dots represent one biological replicate. (C) SPRINT measured constitutive transcription of Cas13a target as transcription was inhibited at increasing concentrations of rifampicin. Bars indicate mean value, error bars indicate s.d. from the mean. n = 3. (D) Diagram of an enzyme-coupled assay. The enzyme hPNP converts inosine to hypoxanthine, which is detected by the guanine riboswitch xpt/pbuE*6U and triggers the SPRINT signal. (E) The enzyme-coupled assay was used to measure enzymatic activity of hPNP. Concentration of inosine was 1 mM, hPNP was added to an activity of 10 mU/μl, concentration of Immucillin-H was 10 μM. Bars indicate mean value, error bars indicate s.d. from the mean. n = 3. p-value was calculated using a two-tailed t-test. (F) The enzyme-coupled assay was used to measure concentration-dependent substrate conversion. hPNP was added to an activity of 1 mU/μl. Bars indicate mean value, error bars indicate s.d. from the mean. n = 3.