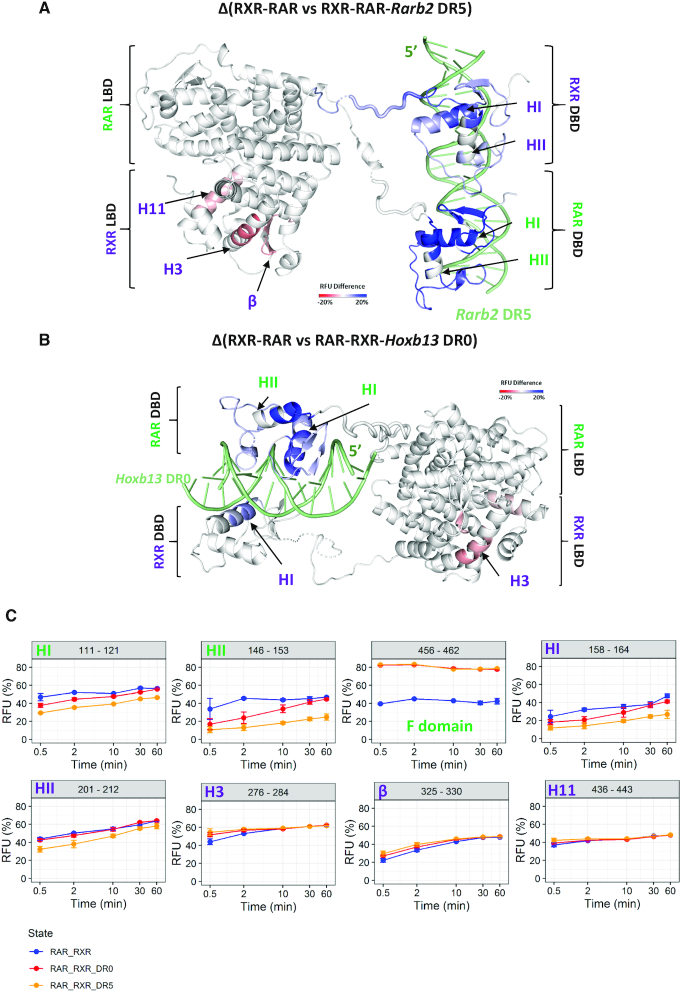
Figure 6.
Conformational dynamics of RXR–RAR upon DNA binding. Export of RFU differences on RXRA ΔAB-RARA ΔAB heterodimer structure upon Rarb2 DR5 (A) and Hoxb13 DR0 (B) bindings determined by HDX-MS. Export is performed for 0.5 min deuteration experiments. RFU differences are color scaled from red (deprotection) to blue (protection) upon DNA binding (–20% to 20% range of RFU difference), while white regions are non-affected or non-covered. Secondary structures exhibiting statistically validated differences for the magnitude are indicated in green for RAR and in purple for RXR. (C) Deuterium uptake of selected perturbed peptides upon DR0 and DR5 binding plotted as a function of deuteration time. Blue, red and orange curves correspond to RXR–RAR, RAR–RXR–Hoxb13 DR0 and RXR–RAR–Rarb2 DR5 states respectively. The corresponding secondary structures are indicated for each plot.