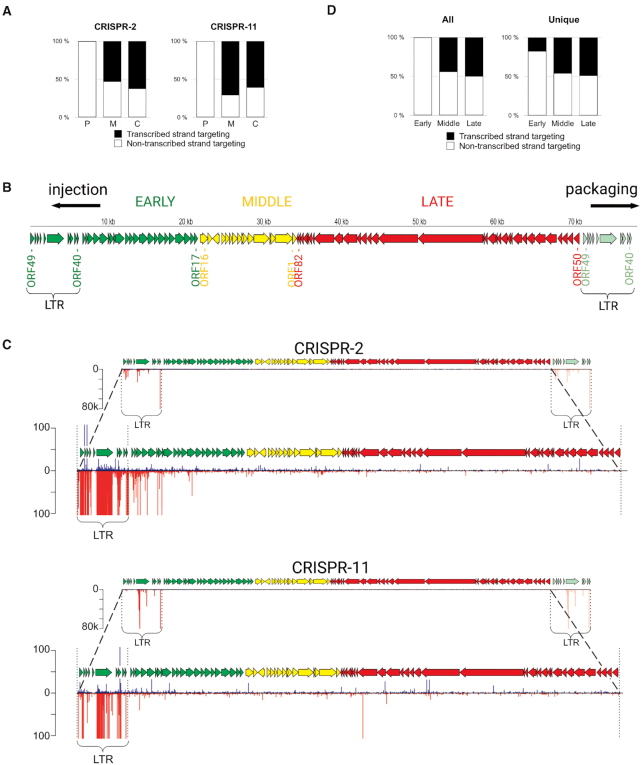
Figure 2.
Analysis of spacers acquired by Type III arrays in phiFa infected cells. (A) Orientation of spacers acquired by the CRISPR-2 and CRISPR-11 arrays. The fraction of spacers parallel to the orientation of genes they are derived from (targeting the transcribed strand) is shown in black, in antiparallel orientation (targeting the non-transcribed strand) – in white. ‘C’ – spacers acquired from the chromosome, ‘M’ – from the megaplasmid, ‘P’ – from the phiFa genome. (B) A revised scheme of phage phiFa genome. Phage genes are shown as colored arrows whose directions match the direction of transcription: green - early genes, yellow - middle, red - late genes. The LTRs and likely directions of phage DNA injection during infection and packaging into virions are shown. (C) Mapping of spacers acquired by CRISPR-2 and CRISPR-11 arrays on the phiFa genome. Spacers whose sequences match the ‘top’ strand of phage DNA (5′-3′ direction) are shown as blue bars, those matching the ‘bottom’ strand – as red bars. The height of a bar reflects the quantity of reads corresponding to a particular spacer. The lower views for each array allow one to see minor spacers (scales indicating the number of reads are indicated on the left). (D) Orientation of phage-originated spacers acquired from phiFa genome regions encoding different temporal classes of genes (early, middle, late). Spacers extracted from reads corresponding to CRISPR-2 and CRISPR-11 arrays were combined for this analysis. Results for all spacers and unique spacers are shown separately. The fractions of spacers targeting the transcribed strands are shown in black, non-transcribed – in white.