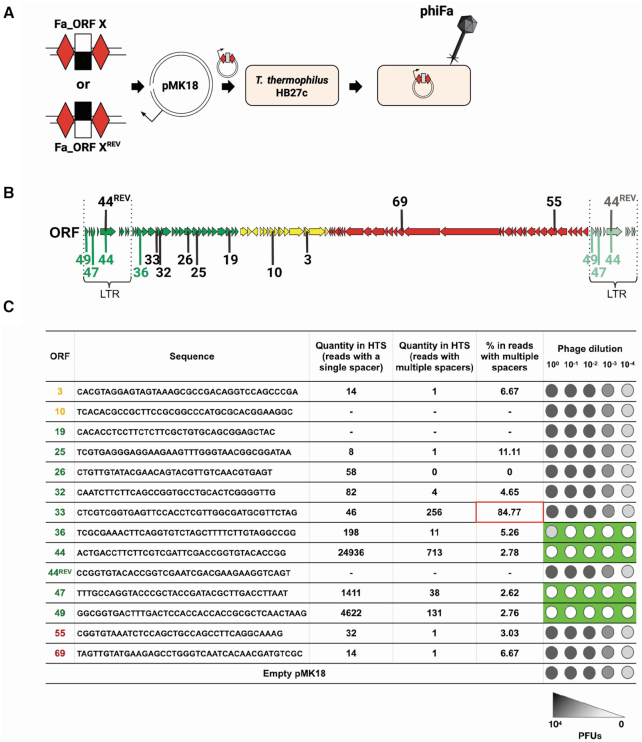
Figure 4.
Type III spacers targeting the phiFa genome outside the “hot" region do not protect from infection. (A) A dsDNA fragment coding a desirable spacer sequence targeting transcribed or non-transcribed strand of phage genes (white/black rectangle) flanked by Type III repeats (red rhombi) is inserted into the pMK18 plasmid. T. thermophilus HB27c cells are transformed with mini-array bearing plasmid and tested for ability to withstand phiFa infection. (B) A scheme of the phiFa genome with protospacers matching spacers tested in plasmid-borne mini-arrays is presented. Spacers are numbered according to phage gene they are derived from. Spacers matching protospacers marked with green color protected cells from infection, spacers matching protospacers marked in black – did not. (C) Sequences of spacers targeting indicated phiFa genes that were chosen to check for phage protection in a plasmid-based assay. The ORFs from which spacers derived are color-coded to show the expression classes (panel B). Quantities of Illumina reads corresponding to each spacer in expanded arrays of cultures after phiFa infection (see Figure 2C) are shown separately for reads containing one or multiple acquired spacers. Spacer 33 targeting an early gene outside the LTR fails to protect the cells when placed on a plasmid-borne mini-array. This spacer was found together with a spacer targeting the LTR-located gene 40, which while not tested separately, is likely to be protective and was the most abundant of all acquired spacers (found in 138 494 reads). The ability of plasmids bearing mini-arrays with these spacers to protect against phiFa infection is schematically presented in the rightmost column (primary data are shown in Supplementary Figure S7).