Abstract
Penile cancer is a rare malignancy with high EGFR expression. In 52 patients, we identified that high EGFR expression was associated with poor tumor differentiation and advanced stage, whereas there was no association of these clinical factors with ERCC1 or TS expression. We identified no KRAS mutations and relatively low expression of ERCC1 and TS compared with other squamous malignancies, which could inform future studies of chemotherapy and targeted therapy.
Objective:
To describe the expression of tissue epidermal growth factor receptor (EGFR), excision-repair cross-complementation group 1 protein (ERCC1), and thymidylate synthase (TS) in patients with penile cancer and explore their association with stage and outcome.
Methods:
A total of 52 patients with penile squamous cell cancer who were treated at the University of Southern California from 1995 to 2010 were identified. Paraffin-embedded tissue underwent mRNA quantitation and immunohistochemistry for expression of EGFR, ERCC1, and TS. KRAS mutations were evaluated using polymerase chain reaction–based sequencing.
Results:
EGFR overexpression was common by mRNA (median, 5.09; range, 1.92-104.5) and immunohistochemistry. EGFR expression > 7 was associated with advanced stage and poor differentiation (P = .01 and .034 respectively) but not with survival in multivariate analysis. ERCC1 mRNA expression was a median of 0.65 (range, 0.21-1.87). TS expression was a median of 1.88 (range, 0.54-6.47). ERCC1 and TS expression were not associated with grade, stage, or survival. There were no KRAS mutations identified. A total of 17 men received chemotherapy; 8 (47%) had an objective response, including 1 with a pathologic complete response. There was a trend for lower expression of EGFR corresponding to a higher likelihood of response (response rate [RR]) to chemotherapy: 67% RR in EGFR mRNA < 7 versus 33% RR in EGFR > 7 (P = .31).
Conclusions:
High expression of EGFR mRNA in squamous cell carcinoma of the penis is associated with advanced stage and poor differentiation, but not survival. In our small heterogeneous subset, molecular marker expression did not show a correlation with the likelihood of chemotherapy response. A prospective evaluation of the role of the EGFR pathway and its regulatory environment in penile cancer is warranted. Given the rarity of this cancer, collaborative prospective cohort evaluations and trials need to be encouraged.
Keywords: Chemotherapy response, DNA repair, Growth factor receptor, Squamous cell carcinoma, Targeted therapy
Introduction
Although squamous cell carcinoma of the penis (PSCC) represents up to 10% of male cancers in Asia and South America, only 1250 new cases are diagnosed each year in the United States and approximately 500 cases are diagnosed each year in the United Kingdom.1 Because of the limited case numbers, it has been difficult to collect cohorts large enough to facilitate examination of the molecular characteristics of PSCC and explore their relationship with clinical outcomes. P53 expression reportedly is associated with poorer outcomes for patients with stage T1.2 However, there is a deficit of data with regard to other molecular markers that are prognostic in other solid tumors and may influence response to systemic therapy, such as excision-repair cross-complementation group 1 protein (ERCC1) and thymidylate synthase (TS).
Frequent overexpression of the epidermal growth factor receptor (EGFR) has been documented in PSCC series,3 although the clinical implications have not been clarified. In head and neck squamous cancers, higher EGFR expression is associated with a higher risk of late relapse, as well as a reduced disease-free and overall survival.4 In vulvar cancer, EGFR overexpression similarly is associated with decreased survival.5 Preliminary reports have suggested that PSCC is responsive to therapies that inhibit EGFR.6 In other malignancies, the ability of EGFR expression by immunohistochemistry (IHC) to predict response has been inconsistent. Alternative techniques, such as fluorescence in situ hybridization, detected gene copy number, and the presence of EGFR or KRAS mutations has been associated with response.7 The clinicopathologic correlates and frequency of EGFR and KRAS mutations in PSCC have not been delineated, but they have potential as prognostic or predictive markers for EGFR-targeted therapy.
The Los Angeles County University of Southern California (USC) medical center cares for a unique population of underserved indigent and “working poor” patients, and has treated a large number of patients with penile cancer. We undertook a retrospective review of all identified patients treated at the Los Angeles County USC and USC Norris Cancer Center between 1995 and 2010 for whom tissue was available for testing of molecular correlates. The goal of the study was to describe the expression of ERCC1, EGFR, and TS in patients with penile cancer and correlate expression levels with clinical and pathologic characteristics and response to therapy.
Materials and Methods
With institutional review board approval (HS-09-00363), patients with PSCC were identified by searching pathology databases. A total of 74 patients were initially identified; 20 did not have tissue available and 2 did not have follow-up available, leaving 52 patients for the study population. This represents an earlier cohort compared with the full clinical cohort published by our institution.8 Charts were reviewed for clinical information, and survival was systematically ascertained using the cancer registries at each center.
Tissue blocks were selected by an experienced pathologist (YM) and sectioned in 10-μm sections for laser-captured microdissection of tumor tissue and real-time polymerase chain reaction (PCR) for mRNA expression by Response Genetics, Inc, Los Angeles, California (KD), a Clinical Laboratory Improvement Amendments–certified laboratory. The methodology of extracting RNA and DNA from paraffin-embedded specimens has been described,9 and patent is pending. Quantitation of RNA was performed using a real-time, fluorescence-based PCR detection method (ABI Prism 7700 Sequence Detection System, Thermo Fisher Scientific, Waltham, MA; TaqMan, Applied Biosystems, Foster City, CA). The output is expression of the gene of interest relative to an internal control gene, β-actin. The DNA extracted from the tumor tissue was added to a 15 μL PCR reaction containing mutation-specific primer/probes, dNTPs, and TaqMan reagents. PCR reactions were run on an ABI Prism 7900HT for 42 cycles with concentrations of reagents and temperatures according to the manufacturer’s recommendations. Primer and probes specific for KRAS mutations were purchased from ABI or Sigma (St Louis, MO). Mutations of interest included any of 6 mutations in codon 12 and a single mutation in codon 13, which are known to result in amino acid substitutions. These mutations are as follows: codon 12 (GGT>GAT), (GGT>GCT), (GGT>GTT), (GGT>AGT), (GGT>CGT), (GGT>TGT), and codon 13 (GGC>GAC). A no template control and extraction control were used as negative controls, and standard positive control was composed of synthetic oligonucleotides mutated for the targeted position.
When there was enough tissue available, additional sections were prepared for IHC with standard deparaffinization and antigen retrieval procedures. Primary antibodies for EGFR and ERCC1 were obtained from AbCam (Cambridge, UK); these were incubated overnight at 3°C and developed using the DAB system (DAKO, Carpinteria, CA). For EGFR, nuclei were lightly counterstained with hematoxylin; for ERCC1, to optimize visualization of nuclear antibody staining, no counterstaining was performed. The intensity of IHC staining was graded by pathologist YM as 0, 1+, or 2+. Normal skin samples initially were used to titrate the antibody concentration to optimize the IHC protocol and later served as positive controls. Most samples were noted to have internal controls, with normal skin next to sections of squamous cancer (Figure 1).
Figure 1.
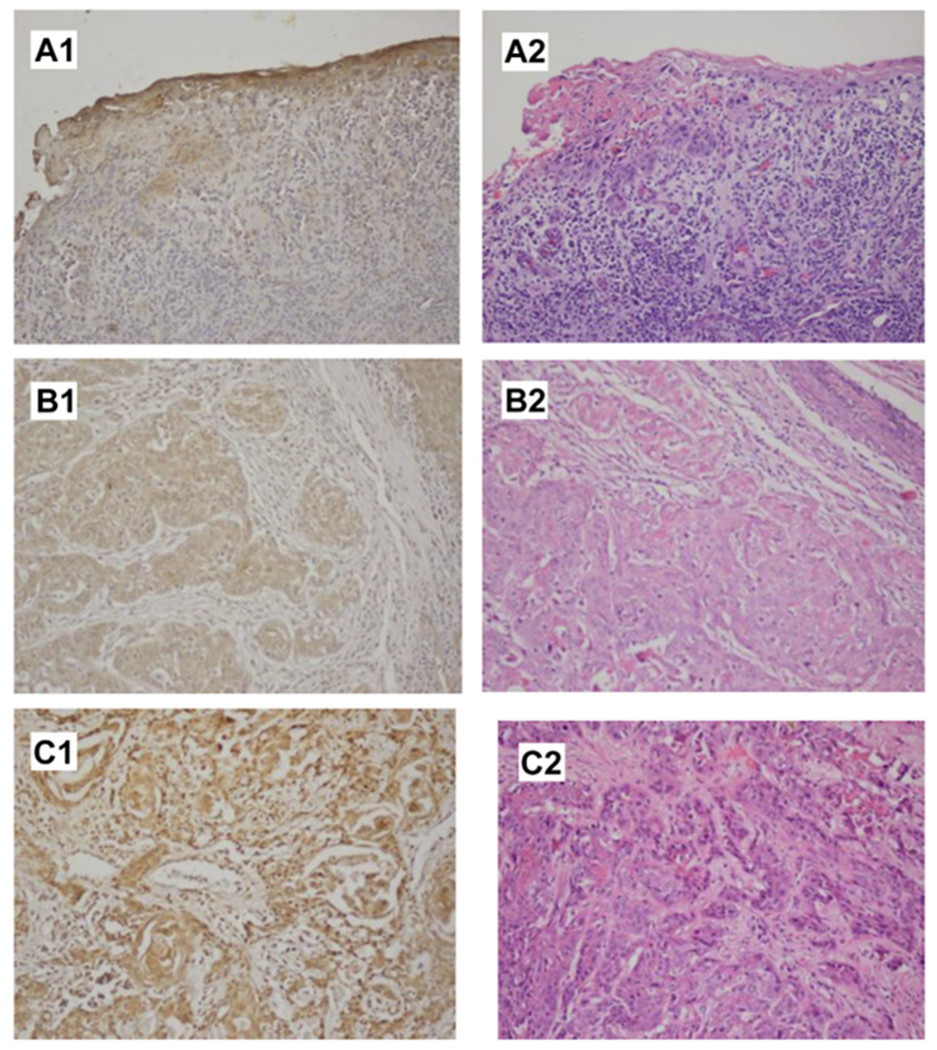
Examples of EGFR IHC in Penile Cancer Specimens. (A1), Negative EGFR IHC in the Tumor Cells (Note the + Internal Control in the Epithelium) With (A2) Showing the Same Area of Tumor With Hematoxylin–Eosin Staining. (B1), EGFR Staining Graded as 1 +, With (B2) Showing the Corresponding Hematoxylin–Eosin Section. (C1), EGFR Staining Graded as 2+, With (C2) Showing the Corresponding Hematoxylin–Eosin Section
Statistical software package SAS Version 9.2 (SAS Institute Inc, Cary, NC) was used for all of the analyses in this study. Pearson’s chi-square or Fisher exact test was used to examine the association between categoric demographic and clinical variables. Wilcoxon rank-sum test was used to test differences in not normally distributed continuous variables between groups or subgroups. Time to overall survival was calculated from the date of diagnosis to the date of death (from any cause) or was censored at the date of last follow-up if the patient was still alive at that time. Kaplan–Meier plots were used to estimate the probabilities of overall survival for every year since diagnosis.10 The log-rank tests were used to compare the differences in survival between dichotomous molecular biomarker RNA expression subgroups, which were based on the cutoff at the median or 75 th percentile for RNA expression level of each biomarker in the dataset. All P values reported are 2 sided.
Results
Baseline and demographic characteristics of the study population are summarized in Table 1. The median follow-up is 2.3 years (longest, 16.8 years). At presentation, 6 patients had stage Tis, 10 patients had T1, 23 patients had T2, 12 patients had T3, and 19 patients had pathologic documentation of lymph node involvement (37%). A total of 31 men had undergone partial penectomy, 11 men had undergone total penectomy, 5 men had undergone organpreserving surgery, and 6 men had undergone an unknown type of surgery; 2 men received pelvic radiation.
Table 1.
Baseline and Demographic Characteristics of the Study Population
| Characteristic | Number (%) |
|---|---|
| Ethnicity | |
| Hispanic | 32 (61) |
| White | 8(15) |
| Asian | 3(6) |
| Black | 3(6) |
| Unknown | 6(12) |
| Age, years | Median 52 (range, 23-80) |
| T stage | |
| Tis | 6(12) |
| T1 | 10 (19) |
| T2 | 23 (44) |
| T3 | 12 (23) |
| T4 | 1 (2) |
| Lymphovascular invasion | |
| Yes | 10 (19) |
| No | 42 (81) |
| LN involvement | |
| Yes | 19 (37) |
| No | 33 (63) |
| Differentiation | |
| Well | 17 (33) |
| Moderate | 24 (46) |
| Poor | 8(15) |
| Unknown | 3(6) |
Abbreviation: LN = lymph node.
No KRAS mutations were identified of 41 samples from which enough DNA was extracted to successfully be tested. Data regarding KRAS mutations and gene expression in our study cohort relative to other cancer cohorts are presented in Table 2.11–27 For the mRNA, EGFR had the highest relative expression (median, 5.09; range, 1.92-104.5), followed by TS (median, 1.88; range, 0.54-6.47), whereas ERCC1 expression was lower (median, 0.65; range, 0.21-1.87). Several samples did not have successful amplification meeting quality-control standards for reporting: 4 for EGFR, 7 for ERCC1, and 7 for TS. Relationships among EGFR, ERCC1, and TS mRNA expression with tumor grade and stage are summarized in Table 3. Higher EGFR mRNA levels were significantly associated with higher stage and poor differentiation (median 9.5 compared with 4.4 for moderate/well-differentiated tumors) on continuous (P = .03 by Mann–Whitney) and cut-point analysis using ≥ 7 (2-sided P = .03 by Fisher exact test). There was no significant correlation for ERCC1 or TS with grade or stage. Fifteen men received systemic chemotherapy, either neoadjuvant or for metastatic disease; all 15 received platinum (cisplatin = 13, carboplatin 1, oxaliplatin 1) partnered with taxane (11) with or without ifosfamide (9), bleomycin (2), or gemcitabine (2). Six men (40%) had progression of disease as their best response, whereas 8 had stable disease and partial or complete response (60%). Tumor expression levels of EGFR, ERCC1, and TS were not associated with chemotherapy response.
Table 2.
Rate of KRAS Mutations and Expression of EGFR, ERCC1, and TS in Our Cohort and in Cohorts Including Other Solid Tumors With Squamous Histology
| Disease and References | KRAS Mutation Presence (%) | Median RNA Expression (Range) or IHC Staining (0-2+) | ||
|---|---|---|---|---|
| EGFR | ERCC1 | TS | ||
| Cervical cancer11–14 | 3/47 (6.3%) | 35.3% (+IHC) | 88.4% (+IHC) | 79% (≥2+ IHC) |
| Esophageal cancer15–17 | 2/36 (5.5%) | 50% (≥2+ IHC) | 1.02 (0.35-13.82) | 2.98 (0.9-14.1) |
| 1.37 (0.51-7.53) | ||||
| Head and neck squamous cancer18–20 | 0%-2.8% | 92% (≥1+ IHC) | 73% (+IHC) | 78% (≥2+) |
| NSCLC21–25 | 36/277 (13%) | 1.98 (0.17-28.27) | 6.7 | 2.17b |
| 88% (≥1+ IHC) | ||||
| Penile cancer26,27 | 0/41a | 5.09 (1.9-104.5)a | 0.65 (0.21-1.87)a | 1.88 (0.54-6.47)a |
| 6/27 (22%) | 100% (≥1+ IHC) | 50% (≥1+ IHC)a | ||
| 91% (≥1+ IHC)a | ||||
Abbreviations: EGFR = epidermal growth factor receptor; ERCC1 = excision-repair cross-complementation group 1 protein; IHC = immunohistochemistry; NSCLC = non–small cell lung cancer.
Indicates the present study findings.
For the squamous subset of NSCLC only.
Table 3.
Relationship Between mRNA Expression of Molecular Markers and Tumor Stage and Grade
| Molecular Markers | Stage/Differentiation | Median mRNA Expression | P Value | Cutpoint | P Value |
|---|---|---|---|---|---|
| EGFR | Tis, T1 | 4.1 | 0 ≥7 | ||
| ≥T2 | 5.7 | .076 | 37% ≥7 | .010 | |
| LN− | 4.7 | 20% ≥7 | |||
| LN+ | 5.8 | .066 | 39% ≥7 | .19 | |
| Well/moderate | 4.4 | 22% ≥7 | |||
| Poor | 9.5 | .012 | 63% ≥7 | .034 | |
| ERCC1 | Tis, T1 | 0.5 | 39% ≥0.7 | ||
| ≥T2 | 0.7 | .29 | 50% ≥0.7 | .53 | |
| LN− | 0.5 | 39% ≥0.7 | |||
| LN+ | 0.9 | .14 | 59% ≥0.7 | .23 | |
| Well/moderate | 0.8 | 53% ≥0.7 | |||
| Poor | 0.5 | .17 | 33% ≥0.7 | .66 | |
| TS | Tis, T1 | 1.3 | 25% ≥2 | ||
| ≥T2 | 2.0 | .91 | 48% ≥2 | .50 | |
| LN− | 1.7 | 46% ≥2 | |||
| LN+ | 1.9 | .84 | 41% ≥2 | 1.00 | |
| Well/moderate | 1.9 | 44% ≥2 | |||
| Poor | 2.6 | .85 | 67% ≥2 | .40 |
Abbreviations: EGFR = epidermal growth factor receptor; ERCC1 = excision-repair cross-complementation group 1 protein; LN = lymph node; TS = thymidylate synthase.
There was no difference in EGFR, ERCC1, or TS mRNA expression in younger (age < 50 years) versus older patients (P = .52 for EGFR, P = .77 for ERCC1, and P = .13 for TS) by Fisher exact test. Race also was not associated with expression of markers when analyzed by Hispanic versus non-Hispanic (P = 1.0 for EGFR, P = .75 ERCC1, and 0.2 for TS). Survival was not associated with T stage (P = .16 for T1/2 vs. T3/4) or differentiation (P = .36 for moderately or well-differentiated compared with poorly differentiated tumors).
Dates of recurrence were not prospectively documented in this retrospective series; therefore, analysis of disease-free survival was not undertaken. Median overall survival was 5.6 years (range, 1 month to 16.8 years). For the 2 men with only 1 month of follow-up, 1 died just after diagnosis of an unrelated cause (lung cancer), and 1 was lost to follow-up 1 month after diagnosis. None of the tested markers were significantly associated with survival; these results are summarized in Table 4 and depicted graphically in Figure 2.
Table 4.
Relationship Between mRNA Expression of Molecular Markers and overall Survival
| Marker RNA Expression | No. of Patients | 5-Year Survival Probability (P ± SE) | Relative Risk | P Value |
|---|---|---|---|---|
| EGFR | ||||
| <7 | 35 | .64 ± 0.10 | 1 | |
| ≥7 | 13 | .63 ± 0.15 | 1.49 | .46 |
| N/A | 4 | – | ||
| ERCC1 | ||||
| <0.7 | 24 | .70 ± 0.10 | 1 | |
| ≥0.7 | 21 | .47 ± 0.21 | 1.08 | .88 |
| N/A | 7 | – | ||
| TS | ||||
| <2 | 24 | .70 ± 0.10 | 1 | |
| ≥2 | 19 | .65 ± 0.15 | 0.98 | .97 |
| N/A | 9 | – |
Abbreviations: EGFR = epidermal growth factor receptor; ERCC1 = excision-repair cross-complementation group 1 protein; N/A = not available; SE = standard error; TS = thymidylate synthase.
Figure 2.
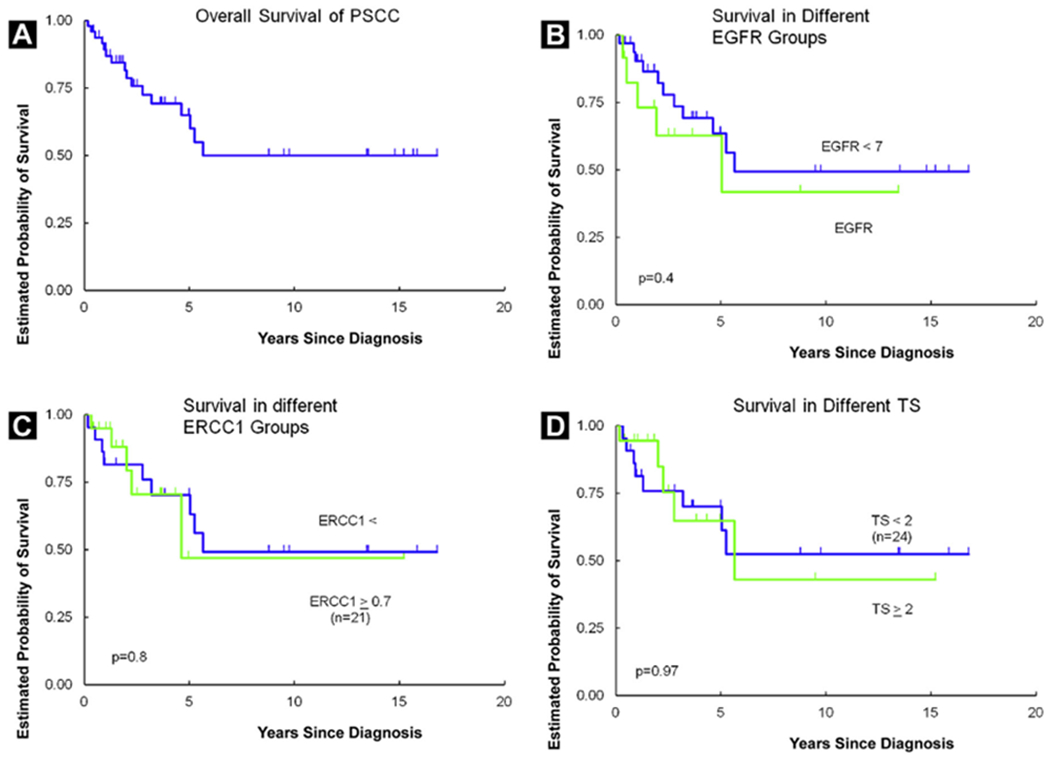
Kaplan–Meier Curves Depicting Overall Survival for the Entire Cohort (A), According to EGFR mRNA (B), according to ERCC1 mRNA (C), and According to TS mRNA (D)
Abbreviations: EGFR = epidermal growth factor receptor; ERCC1 = excision-repair cross-complementation group 1 protein; PSCC = squamous cell carcinoma of the penis; TS = thymidylate synthase.
IHC was performed on a subset of 22 patients for whom there was adequate tissue. For EGFR, 7 of 22 samples had 2+ staining (32%), 13 samples had 1+ staining (59%), and only 2 specimens had no appreciable tumor staining. For ERCC1, 4 of 22 had 2+ staining (18%), 7 had 1+ staining (32%), and 11 had no staining (50%). Low expressing samples by IHC were re-run in a subsequent batch for confirmation. No significant correlation or association was detected between mRNA expression and IHC staining intensity/positivity among those biomarkers in this limited subset. EGFR and ERCC1 IHC and mRNA results are detailed in Supplemental Table 1, in the online version.
Discussion
Penile SCC has a unique profile of EGFR, ERCC1, and TS expression compared with other more common cancers. Our series of 52 patients is unique because we have explored protein and gene expression of EGFR, ERCC1, and TS in patients with penile cancer and their relationship with clinical stage, grade, and clinical outcomes. A significant strength of our study is the ability to use population-based, death certificate–linked cancer registry data to confirm survival status. The major limitation of our study is size, despite the fact that this is one of the largest PSCC cohorts reported. Power calculation suggests we would need approximately 100 patients per group to have 80% power to detect relative differences in the range of 33%. With the small subset given systemic chemotherapy, we did not have adequate power to identify associations between markers and chemotherapy response. Although the genes studied could nevertheless have prognostic value because of biologic influence on tumor behavior, they were primarily selected for their relevance to systemic cytotoxic therapy. Nevertheless, our findings may have some relevance to therapeutic development.
EGFR was selected as a gene of interest given its high expression level, reported prognostic impact in other squamous malignancies,4,5 and potential as a therapeutic target. We found that KRAS mutations were extremely rare in our PSCC population; we saw none in 41 tested specimens despite the use of a Clinical Laboratory Improvement Amendments–certified laboratory. This is in keeping with low rates of KRAS mutations in other squamous malignancies (Table 2) and mirrors another study from China that found only 1 of 94 samples had a KRAS codon 12 mutation, and no BRAF mutations were seen.27 The prevalence of KRAS mutations was higher (6/27 samples) in a series from Spain, and all were noted to be G12D mutations.26 This is likely related to differences in the study populations; the Spanish study did have a higher median age (73 years) compared with our median age of 52 years, and perhaps less ethnic heterogeneity. The low rate of KRAS mutations may portend a high rate of responsiveness to anti-EGFR therapy, because KRAS mutations in patients with colorectal cancer have been associated with resistance.28 Indeed, preliminary reports document clinical responses to anti-EGFR therapy for patients with PSCC.7 A multicenter prospective trial of afatinib for patients with progression after platinum therapy for PSCC will open this year. EGFR expression was high by IHC, mirroring a smaller study of 17 PSCC cases,28 and by mRNA (Table 2). EGFR mRNA expression was associated with advanced stage and poor differentiation, but was not associated with survival or chemotherapy response, although there was a trend toward inferior outcomes in men with higher EGFR expression. Further exploration of this marker in patients receiving systemic therapy would be helpful to further understand the mechanism by which EGFR may affect disease course. There is a lack of concordance between our mRNA and IHC results (Supplemental Table 1, in the online version); this has been noted in the literature29 and is thought to potentially represent post-translational modification events and differential sample composition (IHC is read in tumor tissue, whereas mRNA is frequently obtained from a mix of tumor and normal host tissue).
ERCC1 was selected for study because of the common use of platinum-based chemotherapy for PSCC and the known predictive value it has shown in non–small cell lung cancer, among others. In the International Adjuvant Lung Trial, only those subjects with negative ERCC-1 expression by IHC experienced a survival advantage from cisplatin-based adjuvant chemotherapy; those with tumors expressing 2+ intensity staining in greater than 50% of cells had equivalent survival regardless of assignment to chemotherapy or observation.24 Although low ERCC1 expression by real-time PCR also predicted longer survival with cisplatin-based chemotherapy treatment in patients with metastatic non–small cell lung cancer, it did not necessarily predict response,30 suggesting that the companion chemotherapy agent(s) and other DNA repair enzymes also may be important. We found relatively lower ERCC1 expression in our PSCC cohort, median < 1, than is seen in other solid tumors treated with platinum chemotherapy (Table 2). This finding may underpin the relatively high response rate seen for the ifosfamide/cisplatin/paclitaxel combination chemotherapy used in advanced PSCC.31 The lack of correlation with survival may be related to short follow-up, as well as the small number of patients in our study who received chemotherapy, for whom ERCC1 status would be expected to have the strongest implications.
TS was included because it was shown to have significant prognostic value in cervical cancer,14 a disease with similar biology with links to human papilloma virus (HPV), as well as predictive value for survival after treatment with 5-fluorouracil in colorectal cancer.32 Higher expression of TS has been seen in squamous cancer of the lung, compared with the other histologic variants,25 perhaps accounting for the lower benefit of TS-targeted treatment with pemetrexed in patients with squamous lung cancer compared with the others, especially adenocarcinoma. In our cohort, TS expression was slightly higher than in gastrointestinal tumors, in which it has been reported to have a median expression of 1.36 (Table 2) and in which 5-fluorouracil is a cornerstone of treatment. 5-Fluorouracil has been used for PSCC treatment in small series, with reported response rates of approximately 25% in combination with cisplatin.33 In our series, the lack of correlation between TS expression and survival and the inverse relationship with chemotherapy response may be due to the fact that only 2 patients received 5-fluorouracil treatment, and that was in combination with a platinum agent.
We hypothesized that the underlying cause of penile cancer, and thus marker expression, might be different between younger and older patients. For instance, chronic balanitis related to diabetes or smoking exposure could contribute more to etiology in older patients, whereas HPV could contribute more to etiology in younger patients. However, we did not see any significant differences in EGFR, ERCC1, or TS expression between younger and older patients or by ethnicity stratification. Given the prognostic role of HPV in oral squamous cancer, investigation of the presence of HPV could be important in further understanding differences in disease behavior. Additional analysis, such as HPV testing, will be undertaken to further explore the hypothesis that HPV involvement may be associated with different expression patterns.
Conclusions
Penile cancer is a rare but often aggressive malignancy that can respond to platinum-based chemotherapy. Little is known about the molecular underpinnings and their implications to prognosis and response to therapy. We found relatively low expression of ERCC1 and TS, as well as a paucity of KRAS mutations. Additional exploration of EGFR and ERCC1 as predictive markers for men with PSCC receiving systemic chemotherapy should be undertaken.
Supplementary Material
Clinical Practice Points.
Limited prospective studies have identified therapeutic benefit from platinum chemotherapy and EGFR-targeted antibody therapy in patients with advanced penile cancer.
EGFR overexpression has been shown to be common in penile cancer; we found high expression by both mRNA and IHC, whereas expression of ERCC1 and TS was relatively low. In this series of 52 patients, higher EGFR expression was associated with more advanced tumor stage and poor differentiation. We also found a lack of KRAS mutations, similar to the low rates seen in other squamous malignancies, which provides support to the rationale for testing EGFR-targeted therapy in this disease. In keeping with the prognostic implications of EGFR overexpression, we found a trend toward association of lower EGFR expression with response to chemotherapy, which makes this a marker of interest for prospective study to help stratify patients for different treatment strategies.
Acknowledgments
The authors thank the Medical Women Faculty Association of USC, whose grant provided the primary funding for the project, the USC Norris Comprehensive Cancer Center Core Grant P30 CA014089 for statistical support, and Response Genetics for performing the mRNA expression and KRAS mutation testing.
Footnotes
Disclosure
The authors have stated that they have no conflicts of interest.
Supplemental Data
Supplemental table accompanying this article can be found in the online version at http://dx.doi.org/10.1016/j.clgc.2016.01.013.
References
- 1.Barnholtz-Sloan JS, Maldonado JL, Pow-Sang J, Giuliano AR. Incidence trends in primary malignant penile cancer. Urol Oncol 2007; 25:361–7. [DOI] [PubMed] [Google Scholar]
- 2.Martins AC, Faria SM, Cologna AJ, Suaid HJ, Tucci S Jr. Immunoexpression of p53 protein and proliferating cell nuclear antigen in penile carcinoma. J Urol 2002; 167:89. [PubMed] [Google Scholar]
- 3.Wood L, Lavens N, Gupta R. EGFR over-expression in squamous cell carcinoma of the penis. J Clin Oncol 2008; 26(15S):16029. [Google Scholar]
- 4.Chung CH, Ely K, McGavran L, et al. Increased epidermal growth factor receptor copy number is associated with poor prognosis in head and neck squamous cell carcinomas. J Clin Oncol 2006; 24:4170–6. [DOI] [PubMed] [Google Scholar]
- 5.Growdon WB, Boisvert SL, Akhavanfard S, et al. Decreased survival in EGFR gene amplified vulvar carcinoma. Gynecol Oncol 2008; 111:289. [DOI] [PubMed] [Google Scholar]
- 6.Carthon BC, Pettaway CA, Pagliaro LC. Epidermal growth factor receptor (EGFR) targeted therapy in advanced metastatic squamous cell carcinoma (AMSCC) of the penis: updates and molecular analyses. J Clin Oncol 2010;28(15_suppl), abstr e15022. [Google Scholar]
- 7.Linardou H, Dahabreh IJ, Kanaloupiti D, et al. Assessment of somatic KRAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small cell lung cancer and metastatic colorectal cancer. Lancet Oncol 2008; 9:962–7. [DOI] [PubMed] [Google Scholar]
- 8.Jayaratna IS, Mitra AP, Schwartz RL, Dorff TB, Schuckman AK. Clinicopathologic characteristics and outcomes of penile cancer treated at tertiary care centers in the western united states. Clin Genitourin Cancer 2014; 12:138–42. [DOI] [PubMed] [Google Scholar]
- 9.Press MF, Finn RS, Cameron D, et al. Her-2 gene amplification, Her-2 and epidermal growth factor receptor mRNA and protein expression, and lapatinib efficacy in women with metastatic breast cancer. Clin Cancer Res 2008; 14:7861–70. [DOI] [PubMed] [Google Scholar]
- 10.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc 1958; 53:457–81. [Google Scholar]
- 11.Pappa KI, Choleza M, Markaki S, et al. Consistent absence of BRAF mutations in cervical and endometrial cancer despite KRAS mutation status. Gynecol Oncol 2006; 100:596–600. [DOI] [PubMed] [Google Scholar]
- 12.Noordhuis MG, Eijsink JJ, Ten Hoor KA, et al. Expression of epidermal growth factor receptor (EGFR) and activated EGFR predict poor response to (chemo) radiation and survival in cervical cancer. Clin Cancer Res 2009; 15:7389–97. [DOI] [PubMed] [Google Scholar]
- 13.Park JS, Jeon EK, Chun SH, et al. ERCC1 (excision repair cross-complementing group 1) expression as a predictor for response of neoadjuvant chemotherapy for FIGO stage 2B uterine cervix cancer. Gynecol Oncol 2011; 120:275–9. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki M, Tsukagoshi S, Saga Y, Ohwada M, Sato I. Enhanced expression of thymidylate synthase may be ofprognostic importance in advanced cervical cancer. Oncology 1999; 57:50–4. [DOI] [PubMed] [Google Scholar]
- 15.Hanawa M, Suzuki S, Dobashi Y, et al. EGFR protein overexpression and gene amplification in squamous cell carcinomas of the esophagus. Int J Cancer 2006; 118:1173–80. [DOI] [PubMed] [Google Scholar]
- 16.Warnecke-Eberz U, Metzger R, Miyazono F, et al. High specificity ofquantitative excision repair cross-complementing 1 messenger RNA expression for prediction of minor histopathologic response to neoadjuvant radiochemotherapy in esophageal cancer. Clin Cancer Res 2004; 10:2794. [DOI] [PubMed] [Google Scholar]
- 17.Joshi MB, Shirota Y, Danenberg KD, et al. High gene expression of TS1, GSTP1, and ERCC1 are risk factors for survival in patients treated with trimodality therapy for esophageal cancer. Clin Cancer Res 2005; 11:2215–21. [DOI] [PubMed] [Google Scholar]
- 18.Anderson JA, Irish JC, Ngan BY. Prevalence of RAS oncogene mutations in head and neck carcinomas. J Otolaryngol 1992; 21:321–6. [PubMed] [Google Scholar]
- 19.Hiraishi Y, Wada T, Nakatani K, Negoro K, Fujita S. Immunohistochemical expression of EGFR and p-EGFR in oral squamous cell carcinomas. Pathol Oncol Res 2006; 12:87–91. [DOI] [PubMed] [Google Scholar]
- 20.Jun HJ, Ahn MJ, Kim HS, et al. ERCC1 expression as a predictive marker of squamous cell carcinoma of the head and neck treated with cisplatin-based concurrent chemoradiation. Br J Cancer 2008; 99:167–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer. Cancer Res 2004; 64:8919. [DOI] [PubMed] [Google Scholar]
- 22.Mukohara T, Kudoh S, Yamauchi S, et al. Expression of epidermal growth factor receptor (EGFR) and downstream-activated peptides in surgically excised non-small cell lung cancer (NSCLC). Lung Cancer 2003; 41:123–30. [DOI] [PubMed] [Google Scholar]
- 23.Dziadziuszko R, Witta SE, Cappuzzo F, et al. Epidermal growth factor receptor messenger RNA expression, gene dosage, and gefitinib sensitivity in non-small cell lung cancer. Clin Cancer Res 2006; 12:3078. [DOI] [PubMed] [Google Scholar]
- 24.Lord RV, Brabender J, Gandara D, et al. Low ERCC1 expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer. Clin Cancer Res 2002; 8:2286–91. [PubMed] [Google Scholar]
- 25.Ceppi P, Volante M, Saviozzi S, et al. Squamous cell carcinoma of the lung compared with other histotypes shows higher messenger RNA and protein levels for thymidylate synthase. Cancer 2006; 107:1589–96. [DOI] [PubMed] [Google Scholar]
- 26.Valverde CM, Hernandez-Losa J, Ferrandiz-Pulito C, et al. BRAF and KRAS mutations in penile cancer and their correlation with clinical features. J Clin Oncol 2011; 29(suppl7), abstr 221. [Google Scholar]
- 27.Goh HF, Li X, Qiu M, et al. Epidermal growth factor receptor (EGFR)-RAS signaling pathway in penile squamous cell carcinoma. PLoS One 2013; 8, e62175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lavens N, Gupta R, Wood LA. EGFR overexpression in squamous cell carcinoma of the penis. Curr Oncol 2010; 17:4–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pascal LE, True LD, Campbell DS, et al. Correlation of mRNA and protein levels: cell type-specific gene expression of cluster designation antigens in the prostate. BMC Genomics 2008; 9:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 2008; 359:1757–65. [DOI] [PubMed] [Google Scholar]
- 31.Olaussen KA, Dunant A, Fouret P, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med 2007; 355:983–91. [DOI] [PubMed] [Google Scholar]
- 32.Pagliaro LC, Williams DL, Daliani D, et al. Neoadjuvant paclitaxel, ifosfamide, and cisplatin chemotherapy for metastatic penile cancer: a phase II study. J Clin Oncol 2010; 28:3851–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnston PG, Lenz HJ, Leichman CG, et al. Thymidylate synthase gene and protein expression correlate and are associated with response to 5-fluorouracil in human colorectal and gastric tumors. Cancer Res 1995; 55:1407–12. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


