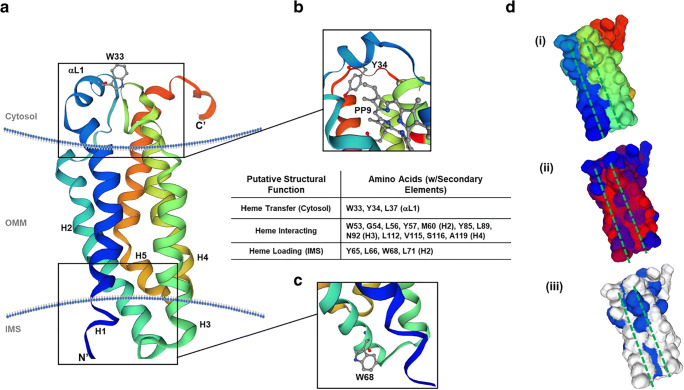
Fig. 7.
Bioinformatics-based modeling of the hTSPO monomer and putative heme-binding capabilities. (a) A ribbon diagram of hTSPO mapped to mouse TSPO structure (PDB: 2mgy.1) with a-helices color coded (blue: Helix 1, blue-green: Helix 2, green: Helix 3, green-yellow: Helix 4, orange-red: Helix 5). The N-terminus (N′) is labeled along with the C-terminus (C′) and conserved tryptophan-33 (W33) in the cytosolic a-loop. (b) immediately below W33 is the proposed porphyrin-binding site identified in crystal structures of Rhodobacter sphaeroides TSPO illustrated by the appearance of protoporphyrin IX (PP9). The conserved a-loop motif of WYXXLXKP, wherein W33, Y34, and L37 of hTSPO were identified as potential heme-interacting residues. Using the HemeBind software and the sequence and modeled structure of hTSPO, we were able to identify amino acids with the potential for heme binding that were not implicated in porphyrin-binding in previous models. Including a motif (WXXLYXXM) in the intermembrane space of the mitochondria that could be the site of heme loading at the close of biosynthesis typified by the presence of W68 in hTSPO at the bottom of helix 2 (c). (d) An examination of the predicted protein surface topology using the same colors as the ribbon structure reveal a groove or trough between helices 1 and 3 (i). This trough has stretches of hydrophobicity (ii, red) and aromaticity (iii, blue) that resemble traits of other heme binding proteins without conserved or strong heme-binding motifs