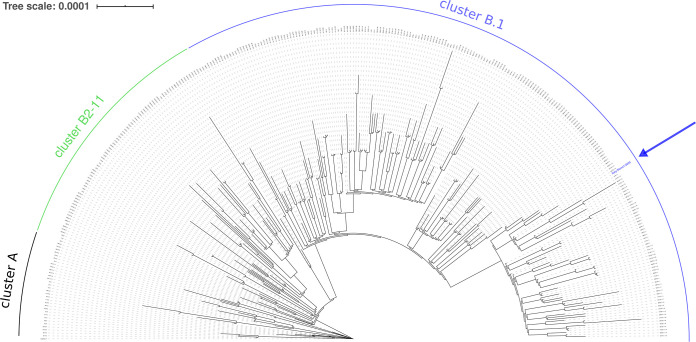
FIG 1.
Phylogenetic tree of SARS-CoV-2 genomes. The tree was generated with Pangolin v1.14 and visualized using Interactive Tree Of Life (iTOL) (8). A total of 322 viral genomes are displayed, including the genomes selected by Pangolin software as representatives for the genetic diversity of SARS-CoV-2. As of 30 July 2020, two major clusters (A and B) were identified. Cluster B was subdivided into 11 clusters (B1 to B11); of those, the most represented is cluster B1 (covered by the blue arch), which comprises most of the lineages identified and sequenced in Europe. Cluster B1.1 is a large subcluster characterized by the mutation of three consecutive nucleotides at position 28881. The blue arrow indicates the position of the Siena-1/2020 isolate in the phylogenetic tree.