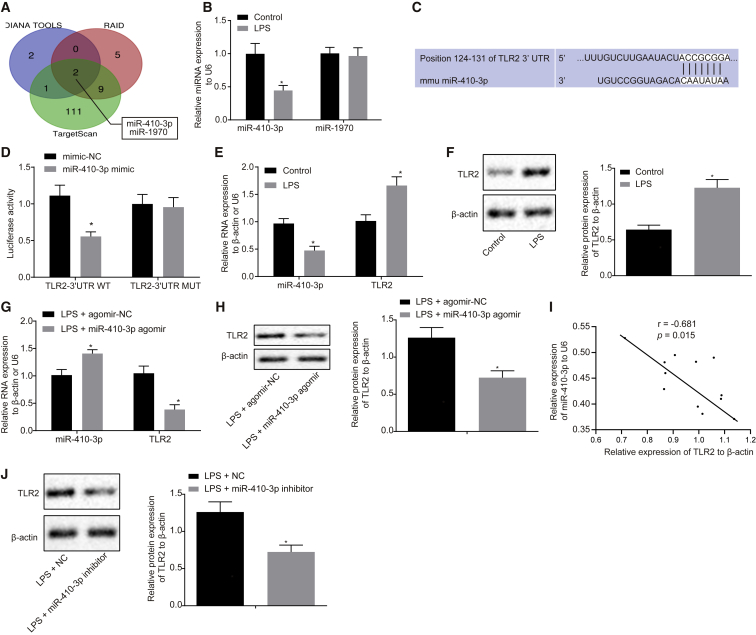
Figure 4.
TLR2 Is a Putative Target of miR-410-3p
(A) Prediction results of miRNAs targeting TLR2 gene by bioinformatics analysis. (B) Expression patterns of miR-410-3p and miR-1970 in myocardial tissues of LPS-treated mice, detected by qRT-PCR, normalized to U6. (C) Putative miR-410-3p binding sites in the 3′ UTR of TLR2 mRNA in the TargetScan website (http://www.targetscan.org/). (D) TLR2 binding to miR-410-3p verified by dual luciferase reporter gene assay in HEK293T cells, n = 3. (E) miR-410-3p expression and TLR2 mRNA expression patterns measured by qRT-PCR in LPS-exposed cardiomyocytes, normalized to U6 and β-actin, respectively. (F) TLR2 protein expression patterns in LPS-exposed cardiomyocytes measured by western blot analysis, normalized to β-actin. (G) miR-410-3p expression and TLR2 mRNA expression patterns measured by qRT-PCR in LPS-exposed cardiomyocytes treated with miR-410-3p agomir, normalized to U6 and β-actin, respectively. (H) Protein expression patterns of TLR2 in LPS-exposed cardiomyocytes after restoration of miR-410-3p, measured by western blot analysis, normalized to β-actin. (I) Correlation analysis of miR-410-3p expression with TLR2 expression in cardiomyocytes of septic mice. (J) TLR2 protein expression patterns in LPS-induced cardiomyocytes treated with miR-410-3p inhibitor measured by western blot analysis, normalized to β-actin. ∗p < 0.05 versus the control (normal mice treated with PBS), mimic-NC (HEK293T cells transfected with mimic NC) or the LPS + agomir-NC group (LPS-exposed cardiomyocytes infected with lentivirus carrying agomir-NC). Measurement data were expressed as mean ± standard deviation. Comparison between two groups was conducted by unpaired t test. n = 12 for mice upon each treatment.