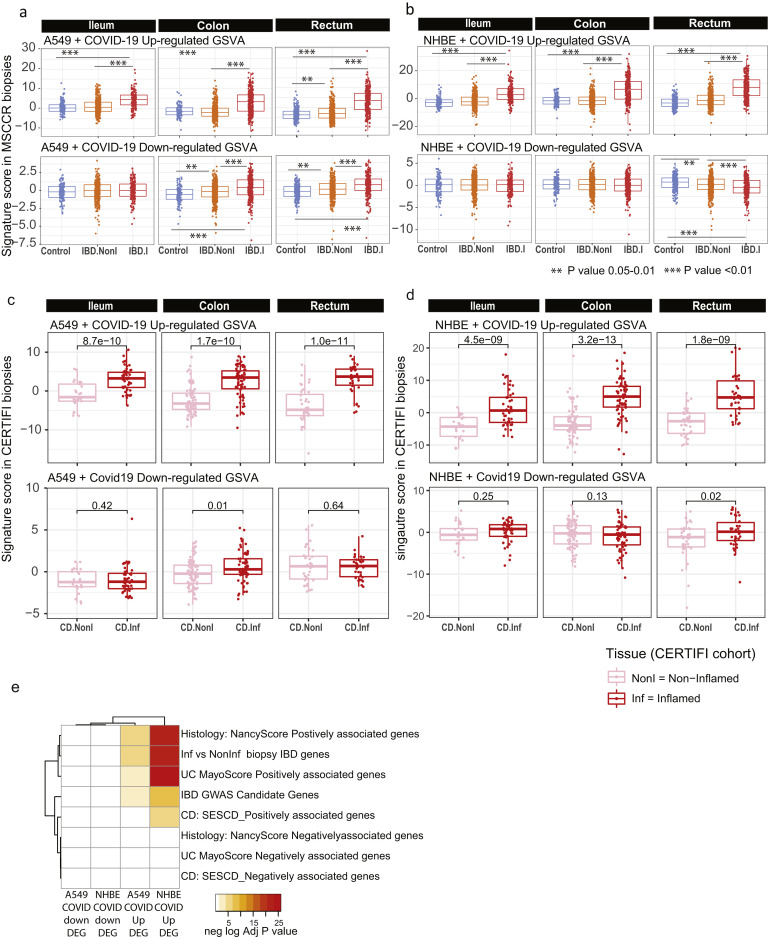
Supplementary Figure 10.
Geneset variation analysis of lung COVID-19 responsive genes as determined in MSCCR and CERTIFI cohort gut biopsies. (A) We evaluated expression of COVID-19–responsive genes as determined in NHBE (A) or A549 (B) lung epithelial cell models22 in the MSCCR biopsy samples using GSVA. Mixed-effect linear models with region, tissue type, and its interaction were used to compare COVID-19 scores among control, noninflamed, and inflamed samples (sample sizes in Supplementary Table 4, ∗∗∗P < .001). (C, D) Two molecular expression signatures reflecting a host’s transcriptional response to SARS-CoV-2 infection were curated from Blanco-Melo et al.22 (C) Transformed lung alveolar cell line (A549) and (D) primary human lung epithelium (NHBE) were profiled following SARS-CoV-2 exposure. Boxplots for the GSVA scores of COVID-19–responsive genes in inflamed and noninflamed biopsies across gut regions for patients in CERTIFI cohort. Mixed-effect models with region and tissue as fixed-effects were used to compare expression between noninflamed and inflamed samples. Sample sizes are presented in Supplementary Table 6. (E) A heatmap summarizing the significance (-log adj P value) for the enrichment of genes up- or down-regulated following lung cell SARS-CoV2 infection in various IBD disease–associated genesets derived from the MSCCR cohort analysis and IBD GWAS genes.