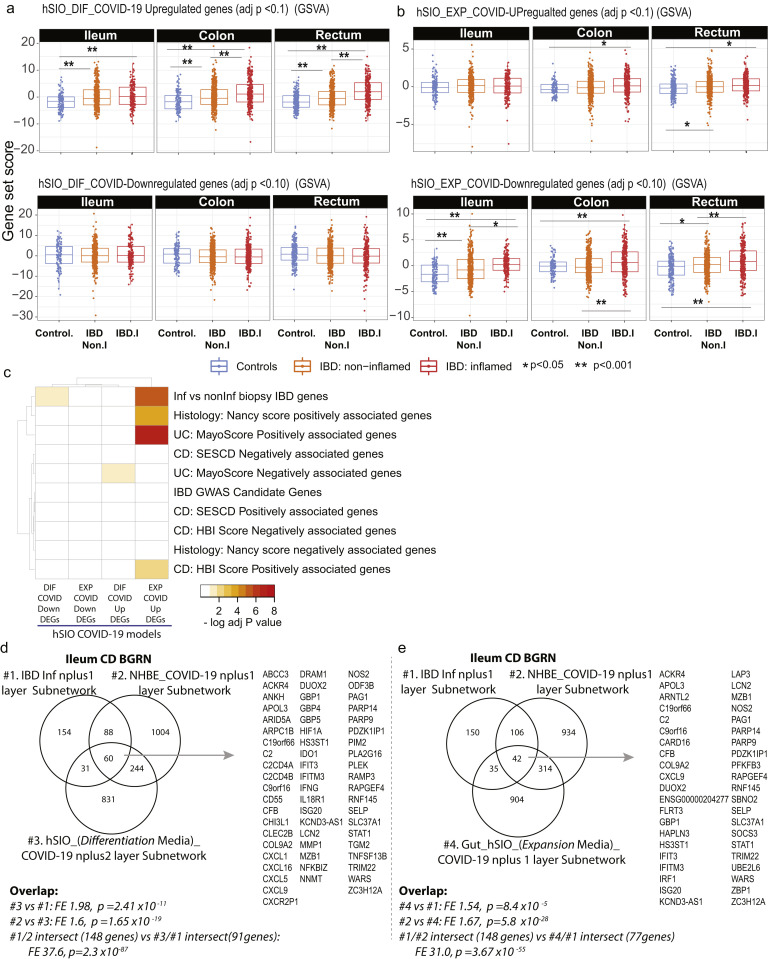
Supplementary Figure 13.
Subnetworks associated with COVID-19 response in hSIOs overlaps with NHBE-COVID-19 responsive subnetworks. Two molecular expression signatures reflecting transcriptional responses to SARS-CoV-2 infection in human small intestinal organoids (hSIOs) were curated.23 (A) hSIOs cultured with differentiation media (DIF) contain predominantly enterocytes, goblet cells, and low numbers of enteroendocrine cells. (B) hSIOs grown in Wnt high expansion medium (EXP) consist mainly of stem cells and enterocyte progenitors. We determined expression of the hSIO COVID-19–responsive genes (at false discovery rate <0.1) in the ileum, colon, and rectum MSCCR cohort samples using geneset variation analysis (GSVA) and mixed-effect model with region, tissue type, and its interaction as fixed effects was performed to compare expression between control, noninflamed, and inflamed samples. P values are as indicated. (C) A heatmap summarizing the significance (-log adj P value) for the enrichment of genes up- or down-regulated following hSIO SARS-CoV2 infection in various IBD disease-associated genesets derived from the MSCCR cohort analysis and IBD GWAS genes. (D) A Venn diagram summarizing the overlaps of 3 subnetworks generated using the ileum CD BGRN, from projecting signatures associated with (1) IBD-Inf; (2) NHBE_COVID-19 response; or (3) hSIO (DIF media)-COVID-19 response, allowing 1 or 2 nearest neighboring genes. (E) A Venn diagram summarizing the overlaps of 3 subnetworks generated using the ileum CD BGRN, from projecting signatures associated with (1) IBD-Inf; (2) NHBE_COVID-19 response; or (3) hSIO (EXP media)-COVID-19 response, allowing 1 nearest neighboring gene. The hSIO(DIF)- and hSIO(EXP)- COVID-19 associated subnetworks are found in Supplementary Table 20. The intersecting genes are shown. Note many genes are also identified as KDGs in Figure 6E. The significance of the overlaps of various genesets is presented in each panel.