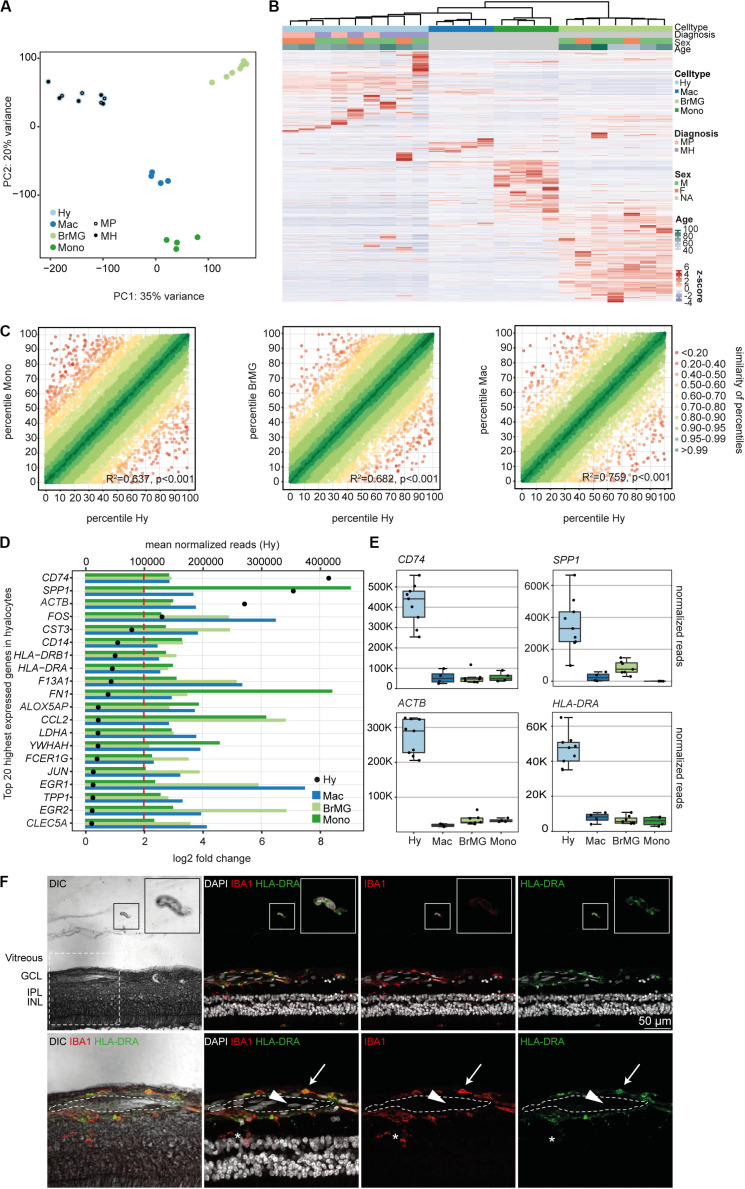
FIGURE 3.
The transcriptional profile of human hyalocytes differs significantly from other myeloid cell populations. (A) Principal component analysis (PCA) plot illustrating the distribution of the four analyzed populations. Hy, hyalocytes; Mac, macrophages; BrMG, brain microglia; Mono, monocytes; MP, macular pucker; and MH, macular hole. (B) Unsupervised heatmap showing the transcriptional profile of hyalocytes, macrophages, brain microglia, and monocytes (all genes of all data sets). Rows and columns are clustered according to similarity of the expression. The color bars on the top of the heatmap reveal cell type (hyalocytes in light blue, macrophages in dark blue, brain microglia in light green, and monocytes in dark green), diagnosis of study subjects (pink for macular pucker, lilac for macular hole), sex (grass-green for male, orange for female), and age (age bar on the right side). Color coding of the transcripts according to the z-score (deviation from a gene’s mean expression in standard deviation units). Hy, hyalocytes; Mac, macrophages; BrMG, brain microglia; Mono, monocytes; MP, macular pucker; MH, macular hole; and NA, not applicable. (C) Graphical presentation of the percentiles of normalized counts to illustrate the similarity between the transcriptomes of monocytes (Mono) and hyalocytes (Hy), brain microglia (BrMG), and hyalocytes or macrophages (Mac) and hyalocytes. The expression of each gene in each cell population was calculated as a percentile (the gene with the highest mean of normalized counts getting the percentile 100, the one with the lowest – the percentile 0). Colors code for the similarity defined as 1 min Δ percentile. The Pearson coefficient R2 quantifies the deviation from the diagonal with the incline 1. The higher R2, the higher the similarity between the two compared cell populations. (D) Top 20 most highly expressed genes in human hyalocytes (Hy). Transcripts are sorted according to their expression level in hyalocytes (mean normalized reads counts, black dot, and upper x-axis). The color bars code for the log2 fold change (lower x-axis) of the upregulated transcripts in hyalocytes in comparison to monocytes (dark green), brain microglia (light green), and macrophages (blue). The dashed red line distinguishes the log2FC of 2, according to which factors are defined as differentially upregulated. (E) Box plots, illustrating the normalized reads count of CD74, SPP1, ACTB, and HLA-DRA in hyalocytes (Hy), macrophages (Mac), brain microglia (BrMG), and monocytes (Mono). (F) Immunohistochemical staining for HLA-DRA and IBA1. A vitreal cell is presented in higher magnification in the upper right corner. Higher magnification of the section within the dashed white square in the lower panel. The lumen of an intraretinal vessel is traced with the white dashed line. The arrow points at a perivascular macrophage, the arrow head to an intraluminal leukocyte, the asterisk is positioned between two microglial cells (positive for IBA1). Nuclei are counterstained with DAPI. DIC, differential interference contrast; GCL, ganglion cell layer; IPL, internal plexiform layer; and INL, inner nuclear layer.