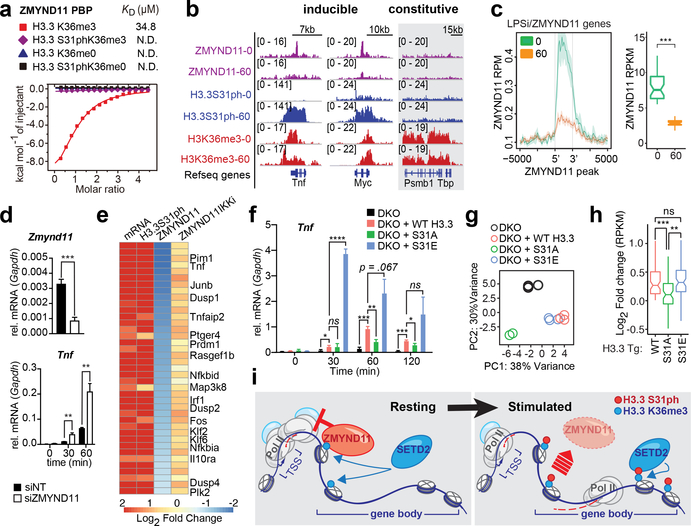
Figure 4: H3.3S31ph ejects transcription repressor ZMYND11 and stimulates transcription.
(A) Isothermal titration calorimetry (ITC) curves of H3.3K36me3, H3.3S31phK36me3, unmodified H3.3 and H3.3S31ph peptides titrated into ZMYND11 PHD-BROMO-PWWP (PBP) domain. (B) ChIPseq tracks of ZMYND11, H3.3S31ph, and H3K36me3 in LPS stimulated BMDM (0 and 60’) for Tnf, Myc, and Tbp. (C) Average gene profiles of ZMYND11 for LPS-induced-ZMYND11-target genes in resting and stimulated BMDM (left) and comparison of ZMYND11 ChIPseq densities before and after LPS stimulation (right). (D) RT-qPCR for Zmynd11 and Tnf after LPS stimulation of BMDM transfected (48 hours before) with siRNA non-target control (NT) and against Zmynd11. (E) Heatmap of LPS-induced-ZMYND11-target genes displaying fold changes between resting and stimulated BMDM for mRNA and ChIPseq densities of H3.3S31ph, ZMYND11 and ZMYND11 after pre-treatment with IKK inhibitors. (F) RT-qPCR for Tnf of H3.3 double knockout (DKO) RAW264.7 cells “rescued” with H3.3 WT, S31A, or S31E transgenes prior to LPS stimulation. (G) Principal component analysis based on expression (RPKM) of all LPS-induced genes. (H) RPKM log2 fold-change of WT, S31A, and S31E compared to DKO for all LPS-induced genes that were “rescued” by WT H3.3 transgene (log2 fold change > 0). (I) Proposed mechanism: During resting conditions the transcriptional repressor ZMYND11 is bound to pre-existing H3.3K36me3 due to constitutive activity of the H3K36me3-specific methyltransferase SETD2. Stimulation-induced H3.3S31ph introduces a steric clash which ejects ZMYND11 and allows for SETD2 to bind, which in turn increases its methyltransferase activity and enables rapid and robust transcription. (G), (H) have three biological replicates per condition. *, p<0.05; **, p<0.005; ***, p<0.0005; ****, p<0.0001; Student’s t-test.