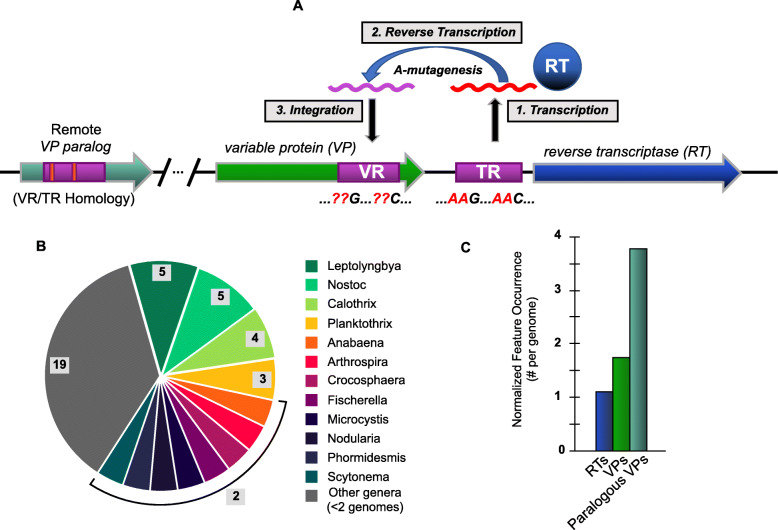
Fig. 1.
Schematic overview of a DGR and their prevalence in cyanobacterial genomes. a Three primary steps in the process of mutagenic homing are shown: 1) conserved template region (TR) in the DGR cassette is transcribed into intermediate, non-coding RNA, which is the substrate for DGR reverse transcriptase (DGR-RT). 2) Template-primed reverse transcription of TR-RNA is highly error-prone at adenines, which thus incorporates random nucleotides at specific positions in the resulting cDNA. 3) The new cDNA molecule is integrated into the variable region (VR) at a fixed locus, resulting in the replacement of a portion of the target gene (~ 100 – 200 bp). Genomic surveys suggest that VRs occur almost exclusively near the 3′ terminus of a target gene. Additional “remote” VP genes (i.e. paralogs) may be found in non-DGR loci throughout the genome, which have detectable TR vs VR homology. b Summary of 52 cyanobacteria genomes known to have DGR components (in Fig. 1a) spanning 31 genera. Genera with ≥2 DGR-containing genomes annotated. c DGR feature occurrence normalized to genome number