Figure 1. Reconstruction of Rhythmic Microbial Oscillations Correlate With and Predict Healthy Versus T2D Incidence in the Large, Regionally Homogeneous Human KORA Cohort.
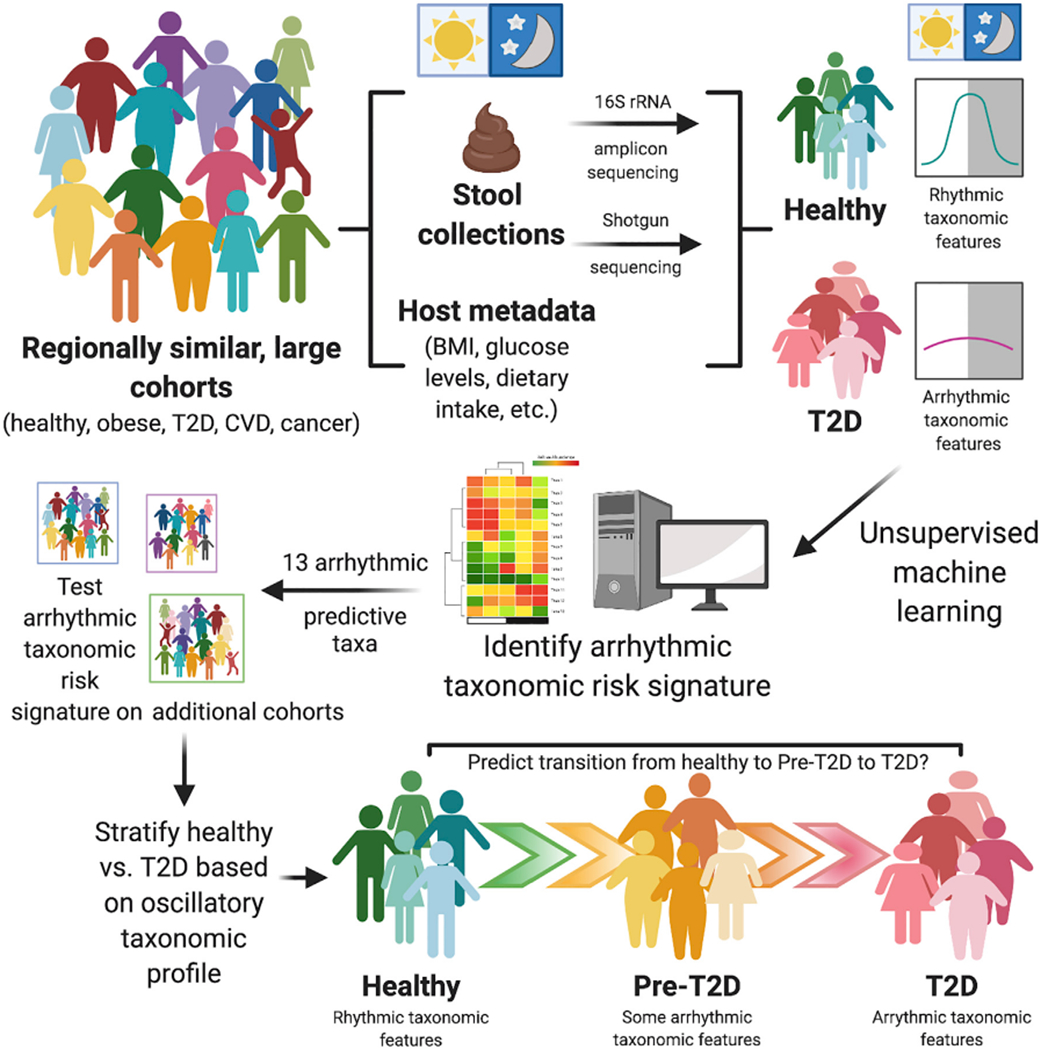
Reitmeier et al. (2020) demonstrate that gut microbiome rhythmicity can be retrospectively reconstructed using fecal collection timestamp data in single stool samples obtained from individual healthy subjects over the course of a day. A number of these microbial oscillations are absent or arrhythmic in Type 2 Diabetes (T2D) subjects. Thirteen taxa were identified as arrhythmic and predictive of T2D in the KORA cohort, which persisted in a 5-year follow-up study and was validated in other large cohorts from similar regions and may eventually allow for stratification of patients into categories of health, pre-T2D, and T2D in patient populations. While these 13 arrhythmic taxa were poor predictors of T2D in stool samples from other large cohorts outside this geographic region, this study provides a framework from which to build upon to incorporate into other timestamped datasets to aid in identifying complex diseases from gut microbiota samples in large heterogeneous cohorts.
