Summary:
The ability to inherit learned information from parents could be evolutionarily beneficial, enabling progeny to better survive dangerous conditions. We discovered that after C. elegans have learned to avoid the pathogenic bacteria Pseudomonas aeruginosa (PA14), they pass this learned behavior on to their progeny, through either the male or female germline, persisting through the fourth generation. Expression of the TGF-β ligand DAF-7 in the ASI sensory neurons correlates with and is required for this transgenerational avoidance behavior. Additionally, the Piwi/Argonaute homolog PRG-1 and its downstream molecular components are required for transgenerational inheritance of both avoidance behavior and ASI daf-7 expression. Animals whose parents have learned to avoid PA14 display a PA14 avoidance-based survival advantage that is also prg-1-dependent, suggesting an adaptive response. Transgenerational epigenetic inheritance of pathogenic learning may optimize progeny decisions to increase survival in fluctuating environmental conditions.
Graphical Abstract
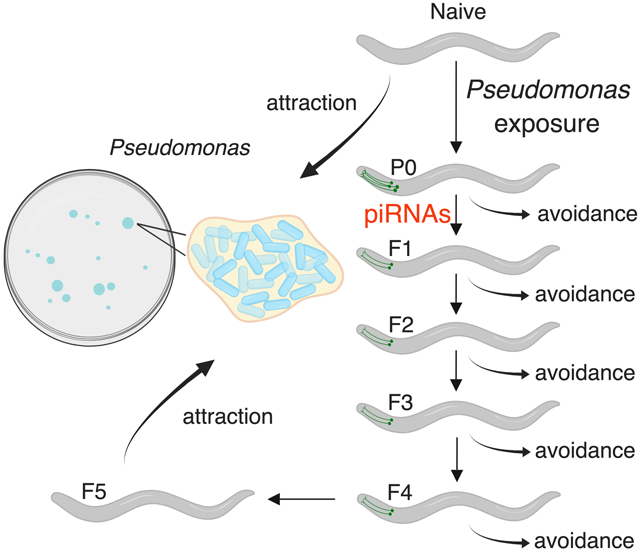
In Brief:
Worms can transmit avoidance behavior to their young for four generations, providing progeny with a mechanism to survive dangerous conditions.
The physical and behavioral characteristics of plants and animals are greatly influenced by the environment. Plasticity in response to environmental changes can be encoded independently of changes to the DNA through a suite of molecular mechanisms collectively known as epigenetics. Epigenetic encoding mechanisms include genome-associated changes, such as DNA and histone modifications and small RNA regulation, and genome-independent changes, including maternal provisioning and microbiome effects. Remarkably, epigenetic changes and their subsequent effects on gene expression are not limited to a single organism’s lifetime, but also can be transmitted across multiple generations in a phenomenon known as transgenerational epigenetic inheritance (TEI). TEI has been shown to play roles in silencing of repetitive elements (such as transposons) (Ghildiyal and Zamore, 2009) and in physiological responses to cellular stresses, such as starvation (Rechavi et al., 2014), heat (Klosin et al., 2017), and osmotic stress (Burton et al., 2017). Despite advances in characterizing TEI and its molecular components (Perez and Lehner, 2019), it is less well understood whether TEI can communicate information encoding the behavior of an animal, and, if so, which epigenetic mechanisms underlie those behaviors.
Caenorhabditis elegans has emerged as a premier organism to study TEI, due to its exceptionally short generation times, large broods, and evolutionarily conserved epigenetic mechanisms that encode and transmit transgenerational information. Moreover, worms do not provide maternal care, and parental microbiomes are not transferred to progeny in laboratory settings—two factors that can confound mechanistic studies of TEI in mammals.
RNA interference (RNAi) was first discovered in C. elegans over two decades ago (Fire et al., 1998), and was later found to be functionally conserved in many species (Grimson et al., 2008), including mammals (Wianny and Zernicka-Goetz, 2000). Subsequently, it was observed that gene silencing induced by double-stranded RNA could be inherited for 3-20+ generations in C. elegans (Vastenhouw et al., 2006). Small interfering RNAs, including exogenous and primary endogenous siRNAs, and Piwi-associated RNAs (piRNAs) trigger an RNA amplification mechanism that guides the production of secondary endo-siRNAs (22G RNAs) (Pak and Fire, 2007) via RNA-dependent RNA polymerases (RdRPs) (Sijen et al., 2001). 22G RNAs are carried by Argonautes to the nucleus (Guang et al., 2008), where they guide the deposition of repressive histone methylation marks that establish gene silencing (Burkhart et al., 2011; Burton et al., 2011; Gu et al., 2012; Mao et al., 2015). Chromatin methylation of H3K9, H3K27, and H3K4 are required for TEI-related phenotypes in worms, flies, and mice (Perez and Lehner, 2019).
The molecular characterization of TEI pathways has provided the framework for understanding how information is encoded and transmitted across several generations, but important questions remain unanswered. For example, while antiviral immunity and physiological stress responses can be transmitted transgenerationally (Rechavi et al., 2011; Vassoler et al., 2013; Tauffenberger and Parker, 2014; Rechavi et al., 2014; Kishimoto et al., 2017; Perez et al., 2017; Burton et al., 2017) it is not known whether complex neurological behaviors, such as learning and memory, can be inherited across generations. Traumatic stress (Gapp et al., 2014) and odor-specific conditioned fear responses (Dias and Ressler, 2014) can be inherited for up to two generations, but the underlying molecular epigenetic mechanisms encoding this behavior remain unknown. Moreover, it is unclear how the currently reported TEI phenomena provide an evolutionary advantage to organisms in the wild. For example, parental mutations of the COMPASS complex of chromatin modifiers extend C. elegans lifespan in both mutant parents and wild-type cross progeny through the third generation (Greer et al., 2011), but it is less clear how these effects might relate to the fitness of wild-type animals in a natural setting. Finally, learned behaviors that might help animals adapt to the environment have not been previously reported to be transgenerationally inherited in C. elegans.
C. elegans inhabit naturally diverse environments (Schulenburg and Félix, 2017), and must constantly discern between nutritious and infectious bacterial food sources (Shtonda and Avery, L., 2006). In fact, ~30% of C. elegans’ natural microbial environment is comprised of Pseudomonae species (Samuel et al., 2016), some of which are pathogens. Although Pseudomonas aeruginosa (PA14) kills C. elegans within hours-days of exposure (Tan et al., 1999), initially, worms are attracted to PA14 – in fact, they prefer it to the non-pathogenic lab E. coli (OP50) – but within hours of exposure, they learn to avoid this pathogen (Zhang et al., 2005). Worms can even form long-term, CREB-dependent memories of the pathogenic experience when exposed as larvae and tested as adults, but this behavior is not transmitted across generations (Jin et al., 2016).
Here, we have found that worms not only learn to avoid PA14, but can also pass on this avoidance behavior to progeny for up to four generations, through TGF-β signaling in sensory neurons and the Piwi/Argonaute small RNA pathway. Worms that inherit the TEI-encoded behavior are able to avoid pathogenic bacteria, providing them with a survival advantage compared to their naïve counterparts. TEI of pathogenic avoidance may allow worms to navigate a complex environment, improving their ability to obtain adequate nutrition while avoiding illness.
Results
Learned pathogenic avoidance is transgenerationally inherited
C. elegans are exposed to and consume a variety of bacterial food sources in their natural environment (Schulenburg and Félix, 2017). Several of these bacteria are pathogens that reduce C. elegans lifespan and progeny production (Tan et al., 1999; Dirksen et al., 2016). Naïve C. elegans initially prefer pathogenic Pseudomonas aeruginosa (PA14) to a laboratory strain of nonpathogenic E. coli (OP50). Upon brief (4h) exposure to PA14, however, worms switch their preference and instead avoid PA14 (Zhang et al., 2005). We wondered whether C. elegans can pass this learned avoidance behavior to their naïve progeny (Fig. 1A). To test this hypothesis, we exposed wild-type L4 (pre-adult) hermaphrodites to PA14 for 4h (Zhang et al., 2005) and bleached the mothers to obtain F1 eggs, but we found that this short exposure does not induce transgenerational effects in the adult progeny (Fig. 1A-B). By contrast, when we exposed the mothers to PA14 for 24h, their adult F1 progeny exhibited avoidance, despite never previously having encountered PA14 (Fig. 1C).
Fig. 1: Pathogenic avoidance can be inherited and is transmitted through both germlines.
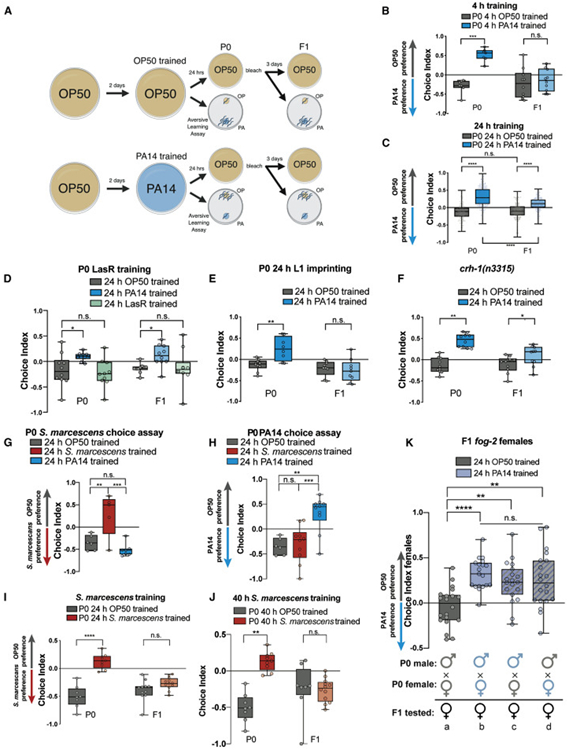
(A) Adult pathogen training protocol. Choice Index = (# of worms on OP50 - # of worms on PA14)/(Total # of worms). (B) 4h of training on PA14 is sufficient to elicit maternal avoidance (P0) of PA14, but is not sufficient for progeny avoidance (F1). At least 3 biological replicates were performed. (C) 24h of training on PA14 induces both pathogen avoidance and transgenerational inheritance of pathogen avoidance (F1); n = 177 replicate assays. (D) 24h of training on avirulent Pseudomonas (LasR) does not induce maternal pathogenic learning or progeny avoidance of PA14. At least 3 biological replicates were performed. (E) L1-imprinted animals (24h) exhibit adult (Day 1) aversion to PA14, but progeny of imprinted mothers do not. At least 3 biological replicates were performed. (F) CREB (crh-1) is not required for maternal pathogenic learning or progeny avoidance of PA14. At least 3 biological replicates were performed. (G) 24h of training on S. marcescens (red) is sufficient to elicit maternal avoidance of S. marcescens. However, 24h of training on PA14 (blue) does not induce maternal avoidance of S. marcescens. 2 biological replicates were performed. (H) PA14 training (blue) induces maternal avoidance of PA14, while S. marcescens (red) training does not induce avoidance of PA14. 24h (I) or 40h (J) of training on S. marcescens is sufficient to elicit maternal (P0) avoidance of S. marcescens, but does not induce progeny avoidance (F1). At least 3 biological replicates were performed. (K) Progeny of (b) PA14-trained mated parents (PA14-trained males x PA14-trained females) inherit pathogen avoidance, while progeny of (a) OP50-trained mated parents (OP50-trained males x OP50-trained females) prefer PA14. Progeny in which only one parent was trained on PA14 ((c) PA14-trained male x OP50-trianed female, or (d) OP50-trained male x PA14-trained female) inherit pathogen avoidance. One-Way ANOVA, Tukey’s multiple comparison test, mean ± SEM. n ≥ 7 choice assay plates containing 50-200 worms. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p < 0.0001, ns = not significant.
A previous study reported that brief (30 min) pre-exposure to the odor of PA14 or a chemical, 2-aminoacetophenone (2AA), can cause temporary (4h) PA14 avoidance (Ooi and Prahlad, 2017). We wondered whether prolonged exposure to odor alone would be sufficient to induce transgenerational memory. However, neither mothers nor progeny of mothers exposed to PA14 odor or 2AA for 24h avoided PA14 (Fig. S1A-B), suggesting that physical contact is required for both pathogenic learning and for the transgenerational inheritance of this behavior. We next asked whether virulence is required for these effects. Prolonged PA14 exposure kills C. elegans within 60h (Tan et al., 1999), cutting short their normal 2-3 week lifespan (Fig S1C). The PA14 quorum-sensing mutant LasR is markedly less virulent to C. elegans such that LasR-exposed worms remain alive after ~70h of exposure, when all of the wild-type PA14-exposed animals have died (Tan et al., 1999). We found that wild-type worms exposed to LasR do not learn to avoid PA14 (Fig. 1D), and progeny of LasR-trained mothers also fail to avoid PA14 (Fig. 1D). Together, these results suggest that pathogenic learning and transgenerational inheritance of pathogen avoidance require both physical contact and infection with PA14.
Early developmental stage larvae (L1) are capable of learning to avoid PA14 (Jin et al., 2016). This “imprinting” results in the maintenance of PA14 avoidance in early adulthood, but is not transmitted to progeny of imprinted mothers (Jin et al., 2016). Because imprinting training is 12h (Jin et al., 2016), we asked whether longer L1 training would cause both adult aversion to PA14 and TEI of pathogen avoidance behavior. Although 24h of larval training was sufficient to elicit parental avoidance of PA14, progeny of parents trained as L1s did not avoid PA14 (Fig. 1E). Furthermore, the transcription factor CREB (crh-1), which is required for L1 imprinting of PA14 avoidance (Jin et al., 2016) and for many forms of long-term memory (Kauffman et al., 2010) is not necessary for transgenerational pathogenic avoidance, further distinguishing these processes (Fig. 1F). Therefore, the mechanism of TEI is distinct from other forms of CREB-dependent, aversive long-term memory, including larval imprinting of PA14 avoidance.
Learned avoidance behavior and TEI is pathogen specific
Pseudomonas aeruginosa exposure induces behavioral responses in mothers and progeny that alter animals’ chemotaxic preferences. We next asked whether these behavioral changes are specific to Pseudomonas exposure and choice, or whether generalized changes occur that alter attraction and avoidance of other known C. elegans pathogens. Like Pseudomonas, naïve worms prefer Serratia marcescens (Sm) to OP50, and training for 24h induces learned Sm avoidance (Pujol et al., 2001) (Fig. 1G). However, PA14-trained worms specifically avoid PA14 and remain attracted to Sm (Fig. 1G), suggesting that the learned avoidance is specific to PA14. Conversely, Sm-trained worms exhibit behavioral responses that are specific to Sm: Sm-trained worms avoid Sm but remain attracted to PA14, similar to naïve animals (Fig. 1H).
Since exposure to S. marcescens induces pathogen-specific changes in learned avoidance behavior, similar to PA14 training, we tested whether that learned avoidance is transmitted to their progeny; however, the progeny of Sm-trained mothers did not avoid Sm (Fig. 1I). S. marcescens kills C. elegans more slowly than does PA14 (Pujol et al., 2001); therefore, we tested whether longer (40h) S. marcescens training is necessary for the transmission of learned avoidance. However, 40h Sm-trained mothers did not pass on the learned avoidance behavior to their progeny, either (Fig. 1J). Together, these results suggest that inheritance of learned avoidance behavior is not a universal response to pathogen exposure, but rather may be specific to Pseudomonas aeruginosa.
Both male and female germlines transmit TEI avoidance information
Transgenerational inheritance has been shown to be transmitted through the male germline in mice (Dias and Ressler, 2014; Gapp et al., 2014); however, the contribution of the mammalian female germline has been less well studied due to technical limitations. Therefore, we took advantage of the tractability of C. elegans (Burton et al., 2017; Klosin et al., 2017) to determine through which germline the learned pathogenic avoidance is transmitted. C. elegans is a hermaphroditic species and typically reproduces through self-fertilization, since males are rare in nature (<0.01% of the population), but males can mate with hermaphrodites to produce cross-progeny, as well (Ward and Carrel, 1979).
To determine the contributions of the male and female germlines in transmission of pathogen avoidance learning, we asked whether sperm or oocytes from trained parents could induce the learned avoidance behavior in the next generation (F1) using fog-2 animals; fog-2 males are normal, but fog-2 hermaphrodites are defective in spermatogenesis, resulting in true females (Schedl and Kimble, 1988). Following PA14 or OP50 exposure, males and females were washed thoroughly to remove bacteria and were mated in the depicted experimental combinations (Fig. 1K) for 24h on OP50-seeded plates containing streptomycin to prevent additional PA14 exposure. Since naïve P0 males already avoid PA14 (Fig. S1D), likely due to sexually dimorphic neuronal gene expression (Hilbert and Kim, 2017) and differences in male mating and foraging behavior (Ryan et al., 2014), we tested the F1 female cross progeny for acquisition of the inherited learned avoidance behavior (Fig. 1K). As expected, the F1 progeny of OP50-trained males x OP50-trained females (a) preferred PA14, similar to naïve animals, while the F1 progeny of PA14-trained males x PA14-trained females (b) exhibited transmission of learned avoidance, demonstrating that mating alone does not diminish the behavior. Interestingly, both PA14-trained males (PA14-trained males x OP50-trained females, (c)) and PA14-trained females (OP50-trained males x PA14-trained females, (d)) transmitted learned avoidance behavior to the next generation, suggesting a role for both sperm and oocytes in intergenerational inheritance of pathogenic avoidance.
TEI of pathogen avoidance persists for four generations
During training, progeny may have been exposed to the bacterial pathogen while still inside the mother, in which case the F1 avoidance effect could not be considered truly transgenerational, but rather should be restricted to one generation. True epigenetic inheritance persists beyond the first generation, and the duration of this persistence often depends on the specific phenotype; therefore, we tested how many generations the learned pathogenic avoidance of PA14 is transmitted. We found that training of mothers (P0) induces pathogenic avoidance in the four subsequent generations, F1-F4 (Fig. 2A-B). Fifth generation (F5) descendants no longer avoid PA14, but instead are attracted to PA14, resuming naïve behavior (Fig. 2A-B).
Fig. 2: Learned pathogenic avoidance lasts through the F4 generation.
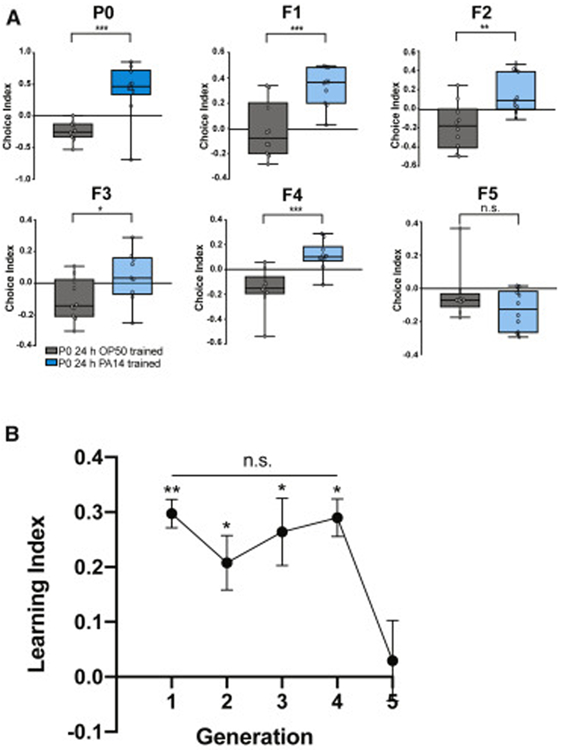
(A) Untrained (naïve) progeny of PA14-trained mothers avoid PA14 from generation F1-F4; the 5th generation returns to normal PA14 attraction. Students t-test, mean ± SEM. n = 10 per generation. At least 3 biological replicates were performed. (B) Learning index (naïve choice index – trained choice index) of generation F1-F5. One-Way ANOVA, Tukey’s multiple comparison test, mean ± SEM. n ≥ 7. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ns = not significant.
Neuronal gene expression changes upon Pseudomonas pathogenic learning and transgenerational inheritance of avoidance
To better understand the molecular mechanisms underlying transgenerational inheritance of pathogen avoidance learning, we compared the transcriptional profiles of wild-type mothers (P0; Fig. S2, Table S1) and progeny (F1; Fig. S3, Table S2), in which the mothers had been treated with OP50 or PA14. Both mothers exposed to PA14 (P0) and their progeny (F1) exhibit a large set of gene expression differences from OP50-trained controls that include immune responses to infection, changes in metabolism, and epigenetic and post-transcriptional regulation of gene expression. Consistent with our observed changes in behavior, the upregulated GO categories (Fig. S3C) were primarily neuronal (ion transport, chemical synapse, neurotransmitter transport, neuropeptide signaling, membrane potential, and behavior); in fact, 65-70% of the genes upregulated by PA14 in P0 and F1 animals, respectively, are expressed in adult neurons (Fig. S3E). These include the TGF-β ligand daf-7, GPCRs, neuropeptides, insulin-like peptides (ins-11, ins-20), MAPK (sek-1), cholinergic signaling, potassium channels and other receptors, and egl-30 Gαq (Table S1). The genes downregulated in the F1 progeny from mothers exposed to PA14 compared to OP50 included several types of metabolism, regulation of gene expression, RNA processing, response to dsRNA, chromatin modifications, and epigenetic gene expression regulation (Fig. S3D, Table S1, S2). Together, these data suggest that in addition to metabolic, immune, and epigenetic changes, the primary upregulated genes in progeny of trained mothers may affect neuron function and behavior.
DAF-7/TGF-β expression and the ASI neuron are required for transgenerational inheritance of avoidance
Sensing and subsequent avoidance of Pseudomonas require the activity of the nervous system (Zhang et al., 2005; Ha et al., 2010; Meisel et al., 2014), and our transcriptional data suggested that neuronal components change upon pathogenic training and learning. Meisel, et al. previously showed that the TGF-β ligand DAF-7 is basally expressed in ASI sensory neurons, but upon PA14 exposure, daf-7 expression increases in the ASI and is induced in the ASJ neurons (Meisel et al., 2014), consistent with our RNA-seq data (Fig. S2). (DAF-7 signaling in the ASJ controls expression of TGF-β signaling in downstream RIM/RIC interneurons, which in turn control reversals through downstream motor neurons (Greer et al., 2008)). We confirmed these observations in mothers exposed to PA14 for 24h (Fig. 3A-D). We then asked whether transgenerational avoidance behavior training induces higher expression of daf-7p::gfp in the ASI or ASJ in progeny of trained mothers. Naïve F1 progeny of PA14-trained mothers maintained a high level of daf-7p::gfp expression in the ASI (Fig. 3E-G), while daf-7p::gfp expression in the ASJ returned to basal levels (Fig. 3F). As we found for pathogenic learning, training with avirulent LasR Pseudomonas did not increase daf-7p::gfp expression in the ASI, nor did it induce expression of daf-7p::gfp in the ASJ of mothers or their progeny (Fig. 3C, G), suggesting that virulence is required for transgenerational daf-7p::gfp expression changes. With 4h of exposure to PA14, daf-7p::gfp expression in the ASJ was induced to higher levels in the progeny of PA14 trained-mothers compared to OP50 trained-mothers (Fig. 3H), but because the choice assay measures an animal’s food preference within a short time frame (<1 h), our daf-7p::gfp expression data suggest that the elevation of basal daf-7 expression in the ASI of progeny, rather than its delayed induction in the ASJ, is likely to mediate transgenerational avoidance behavior. Indeed, genetically-ablated ASI worms avoid PA14 after P0 training, similar to wild-type animals (Fig. 3I), but the F1 progeny of pathogen-trained ASI mutants do not inherit their learned avoidance of PA14 (Fig. 3J), indicating that the ASI is required for pathogen avoidance in the F1s. Moreover, loss of daf-7 causes no defects in the mother’s ability to avoid PA14 or to learn avoidance after 24h of training (Fig. 3K), but abolishes the ability of their F1 progeny to avoid PA14 (Fig. 3L). To more definitively rule out a role for daf-7 in the P0 generation, we used RNAi to knock down daf-7 exclusively in the F1 generation following parental PA14 training. We found that reduction of daf-7 in P0 animals had no effect on P0 learned PA14 avoidance (Fig. 3M). However, when only the F1 progeny of trained mothers were treated with daf-7 RNAi, the avoidance behavior was lost (Fig. 3N), suggesting that daf-7 acts in the F1, rather than the P0, to mediate avoidance.
Fig. 3: DAF-7/TGF-β expression and the ASI neuron are required for transgenerational inheritance of avoidance.
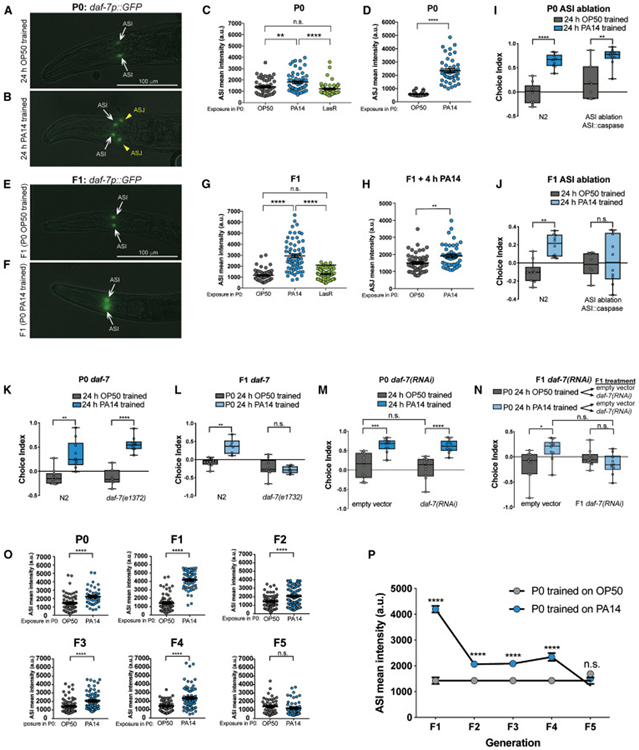
(A) daf-7p::gfp is expressed in the ASI neuron of naïve animals (white arrow). (B, D) PA14 training induces daf-7p::gfp expression in the ASJ neuron (yellow arrow head). Students t-test, mean ± SEM. n ≥ 36 neurons per training condition, 3 biological replicates. (C) PA14 training increases daf-7p::gfp expression in the ASI compared to training with OP50 or LasR. One-Way ANOVA, Tukey’s multiple comparisons test, mean ± SEM. n ≥ 54 neurons per training, 2 biological replicates. (E-G) Naïve progeny of PA14-trained mothers have increased expression of daf-7p::gfp in the ASI compared to progeny of OP50- or LasR-trained progeny. One-Way ANOVA, Tukey’s multiple comparisons test, mean ± SEM. n ≥ 61 neurons per training condition, 3 biological replicates. (H) Progeny (F1) of PA14-trained mothers have higher daf-7p::gfp expression in the ASJ after 4h of PA14 training. Students t-test, mean ± SEM. n ≥ 44 neurons per training condition. (I) The ASI is not required for maternal (P0) pathogenic learning, but is required for progeny (F1) avoidance of PA14 (J). (K) Like wild type (N2), daf-7 mutants (P0) have normal naïve preference and can learn to avoid PA14 after training, but naïve progeny (F1) of daf-7(pk720) PA14-trained progeny do not avoid PA14 (L). (M) PA14 training elicits maternal avoidance of neuron RNAi-sensitive (LC108) worms treated with control vector or daf-7 RNAi bacteria. (N) P0 worms were trained on OP50 or PA14 without any RNAi exposure, then F1 progeny were exposed to control or daf-7 RNAi to test the requirement of daf-7 exclusively in F1s. Progeny of PA14-trained mothers treated with daf-7 RNAi lose the TEI avoidance behavior. One-Way ANOVA, Tukey’s multiple comparison test, mean ± SEM. n ≥ 7 choice assay plates with 50-200 worms. (O, P) PA14 training increases daf-7p::gfp expression in the ASI of progeny of PA14-trained mothers for four generations (F1-F4) before returning to low levels in the 5th generation (F5). Students t-test, mean ± SEM. n ≥ 44 neurons per condition, 2 biological replicates. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p < 0.0001, ns = not significant.
Next, we asked how many generations the increased expression of daf-7 in the ASI persists after pathogenic training. daf-7p::gfp remained elevated in the ASI of progeny of PA14 trained-mothers for four generations, then returned to basal levels in the fifth generation (Fig. 3O-P), similar to the trajectory of transgenerational avoidance (Fig. 2B). Together, our results suggest that daf-7 expression in the ASI neurons of the progeny of PA14-trained mothers is required for transgenerational avoidance behavior, and progeny who inherit PA14 avoidance behavior maintain high ASI daf-7p::gfp expression levels.
COMPASS complex histone modifiers are required for pathogenic learning
To identify possible regulators of epigenetic effects, we investigated the role of candidate TEI regulators. Transient mutation of the COMPASS histone modification complex components induces a transgenerational longevity effect that, like TEI of pathogen avoidance, lasts for four generations (Greer et al., 2011). However, we found that mutants of the PRC1/COMPASS complex H3K4me3 methyltransferases (set-2, wdr-5.1), the H3K4me3 demethylase rbr-2, and mutants of set-32, a putative H3K9me3 methyltransferase that links histone modifications and siRNAs (Woodhouse et al., 2018a), were already defective in either their naïve P0 attraction for PA14, or in aversive pathogenic learning (Fig. 4A-C), so their roles in subsequent transgenerational effects are unclear (Fig. S4A-D, S5A-C). To further investigate possible epigenetic differences, we compared the global H3K4me3 and H3K9me3 methylation levels of OP50 and PA14-treated P0 mothers; although the worms exposed to PA14 are sick and their germlines are diminished, the global methylation level is not obviously altered (Fig. S4H-I), suggesting that global histone methylation is unlikely to be the primary mechanism by which avoidance behavior is inherited.
Fig. 4: COMPASS complex histone modifiers and small RNA regulators are required for pathogenic learning.
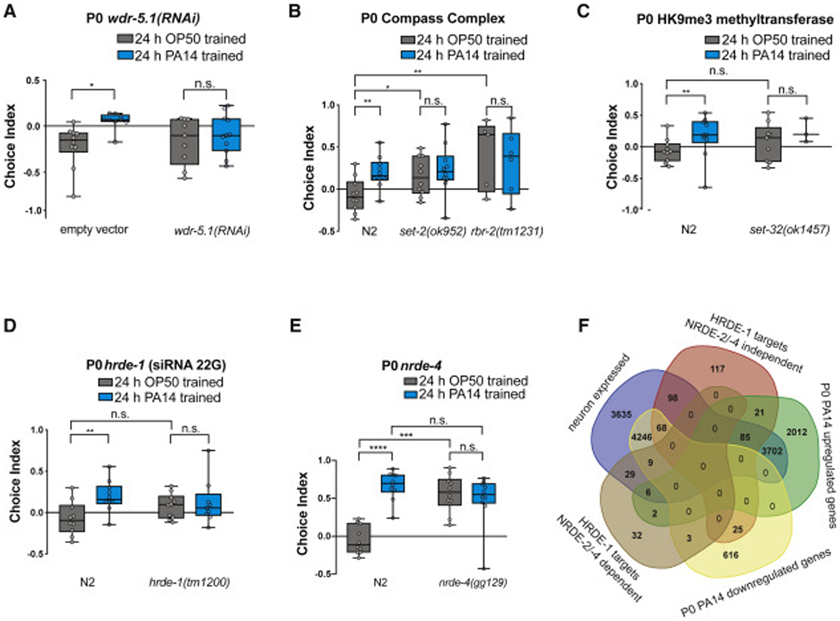
(A) wdr-5.1(RNAi) animals have normal naïve preference, but are defective for pathogenic learning, (B) set-2 and rbr-2 mutants are defective for naïve preference. (C) set-32 mutants have normal naïve preference, but are defective for pathogenic learning. One-Way ANOVA, Tukey’s multiple comparison test, mean ± SEM. n ≥ 7 choice assay plates with 50-200 worms. *p ≤ 0.05, **p ≤ 0.01, ns = not significant. (D) hrde-1(tm1200) mutants have normal naïve preference, but are defective for pathogenic learning. (E) nrde-4(gg129) mutants have high naïve preference and do not appear to learn. One-Way ANOVA, Tukey’s multiple comparison test, mean ± SEM. n ≥ 7 choice assay plates with 50-200 worms. **p ≤ 0.01, ***p ≤ 0.001, ****p < 0.0001, ns = not significant. (F) NRDE2/4-dependent and -independent HRDE-1 targets (Buckley et al., 2012) were examined for overlap with PA14-upregulated and downregulated genes in P0 animals (FDR < 5%), as well as adult neuron expression (Kaletsky et al., 2016).
Small RNA regulators are required for naïve preference and pathogenic learning
Regulators of small RNAs (siRNAs), such as the nuclear argonaute HRDE-1/Ago (Buckley et al., 2012), have also been implicated in TEI (Rechavi et al., 2014). Similar to the histone modification mutants (Fig. 4A-C), hrde-1 mutants had normal attraction to PA14 but were defective for pathogenic aversive learning (Fig. 4D), and thus their transgenerational inheritance of learning could not be tested (Fig. S4E, S5D-E). We also tested nrde-4, which functions downstream of hrde-1 to link siRNAs to H3K9me3 (Burkhart et al., 2011), for its role in TEI. nrde-4 mutants were defective in naïve attraction to PA14 (Fig. 4E, S4F), and thus their transgenerational inheritance of learning could not be tested. Therefore, we instead asked whether previously-identified direct (chIP-seq) HRDE-1 targets (Buckley et al., 2012) overlapped with the differential gene expression changes we found in the P0 mothers (Fig. S2), and we determined the overlap with genes that we previously found to be enriched in neurons (Kaletsky et al., 2016). Strikingly, a large fraction of the PA14-induced genes (3793/5828, or 65%), and NRDE-2/4 independent HRDE-1 targets (251/414, 61%) are neuronal (Fig. 4F), suggesting that HRDE-1 may regulate neuronal genes in response to PA14 exposure.
Exposure to Pseudomonas induces changes in piRNA abundance
Because nrde-4 and hrde-1 have defects in naïve attraction to and learned avoidance of PA14, respectively, we could not easily assess whether these small RNA regulators play a role in transgenic inheritance of learned avoidance. To directly determine whether P0 treatment with PA14 affects small RNA expression as a result of PA14 treatment, we isolated RNA from PA14-treated and control mothers and identified small RNAs (21-26 nts) that are significantly changed upon PA14 treatment (Fig. 5A, Table S3). As expected, several microRNAs, including those previously associated with responses to PA14 (mir-233, let-7, mir-355, mir-75, mir-63, mir-84, mir-241, mir-251, and mir-252) (Kudlow et al., 2012; Liu et al., 2013; Dai et al., 2015; Ren and Ambros, 2015; Ma et al., 2017), were significantly differentially expressed (Fig. 5B). More strikingly, a large group of non-coding, small Piwi-interacting RNAs (piRNAs) were significantly changed upon exposure to PA14 (Fig. 5C, Table S3), confirming a change in piRNA abundance in response to environmental changes (Belicard et al., 2018). The majority of piRNAs were downregulated in response to PA14 exposure (466) compared to those with increased expression (254) (Fig. 5C).
Fig. 5: The PRG-1/Piwi pathway is required for transgenerational inheritance of pathogenic aversive learning.
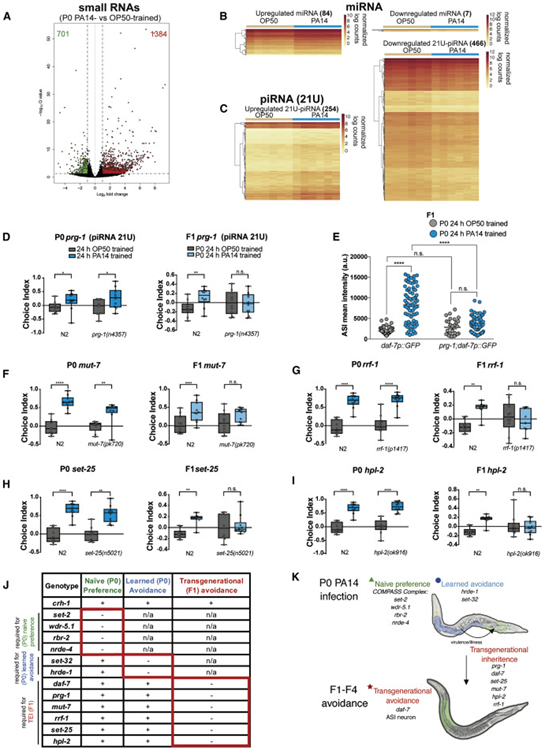
(A) PA14-upregulated (1364, red) and downregulated (701, green) small RNAs were differentially expressed using DESeq2. Volcano plot shown at FDR < 5% and log2 fold change > and < 1. Regularized expression counts from differentially expressed miRNAs (B) and piRNAs (C) in P0 animals. (D) prg-1(n4357) mutants are defective for transgenerational inheritance of pathogenic avoidance. (E) Elevated daf-7p::gfp expression is abrogated in progeny of prg-1 PA14-trained mutants. (F-I) mut-7(pk720), rrf-1(pk1426), set-25(n5021), and hpl-2(ok916) P0s have normal naïve preference and can learn to avoid PA14 after training; however, naïve progeny are defective for inherited avoidance. One-Way ANOVA, Tukey’s multiple comparison test, mean ± SEM. n ≥ 7 choice assay plates with 50-200 worms. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p < 0.0001, ns = not significant. (J, K) Naïve C. elegans prefer PA14, which requires the activity of the COMPASS complex (SET-2, WDR-5.1, and RBR-2) and NRDE-4. After infection by PA14, C. elegans learn to avoid PA14, which requires HRDE-1 and SET-32. piRNA pathway components (PRG-1, MUT-7, RRF-3, RRF-1), and histone modification regulators (SET-25 and HPL-2) are required to transmit transgenerational inheritance of pathogenic avoidance from the P0-F1. daf-7 expression is induced in the ASI neuron, and daf-7 and ASI are required for avoidance in the F1 generation. daf-7 levels in the ASI may set the avoidance response ability of that generation, rendering the animals “primed” for increased expression of daf-7 in the ASJ and subsequent avoidance upon PA14 encounter. (Yellow = neurons, Blue = intestine, Green = germline.). Table: genes are characterized by their requirement for normal naïve preference for PA14 (green), Learned avoidance (blue), and Transgenerational Epigenetic Inheritance (red).
PRG-1/Piwi is required for TEI of pathogenic avoidance and induced daf-7 ASI expression in the progeny of trained mothers
The differential expression of piRNAs upon PA14 treatment suggested that Piwi may play a role in transgenerational inheritance of pathogenic learning. PRG-1 is the C. elegans ortholog of the Piwi Argonaute, and has been implicated in transgenerational inheritance of transposon silencing (Brennecke et al., 2008; Grentzinger et al., 2012) and germline immortality (Simon et al., 2014; Heestand et al., 2018), but has not previously been implicated in response to PA14. Indeed, mutants of prg-1/Piwi display normal naïve choice preference and normal pathogenic learning after PA14 training (Fig. 5D). However, the progeny of trained prg-1/Piwi mothers were defective in their avoidance of PA14 (Fig. 5D, Fig. S4G). These data suggest that PRG-1/Piwi is specifically required for the inheritance of learned pathogenic avoidance behavior.
To test whether PRG-1 activity is required for the regulation of DAF-7, the TGF-β ligand whose expression correlates with avoidance behavior in post-P0 generations and is required for TEI of pathogenic learning (Fig. 3), we examined the expression level of daf-7p::gfp in progeny of PA14-trained prg-1 mutants. While control daf-7p::gfp F1 animals exhibited increased GFP fluorescence in the ASI neuron after P0 training with PA14 compared to OP50 exposure, loss of prg-1 completely abrogated this fluorescence change (Fig. 5E). This result suggests that Piwi/PRG-1 Argonaute activity is required for the induction of daf-7 expression in the ASI neurons of F1 animals, which in turn is required for transgenerational pathogenic avoidance behavior.
The PRG-1/Piwi 22G siRNA pathway is required for transgenerational inheritance of pathogenic aversive learning
Next, we tested components of the nuclear RNAi pathway downstream of prg-1 that have been implicated in transgenerational epigenetic inheritance. RNA-dependent RNA polymerases (RdRPs) amplify primary siRNAs and are essential for the production of secondary siRNAs that execute TEI. To test the contribution of RdRPs to TEI of aversive learning, we tested mut-7, an RNAse D homolog and part of the Mutator complex (Ketting et al., 1999) and rrf-1, an RdRP required for 22G siRNA biogenesis (Aoki et al., 2007). Like prg-1 mutants, mut-7 and rrf-1 have normal P0 naïve attraction to and learned avoidance of PA14, but are specifically defective in F1 avoidance of PA14 (Fig. 5F, G), suggesting that they are part of the prg-1-mediated inheritance mechanism of learned avoidance.
Once secondary siRNAs are made, they are able to direct chromatin modifications via histone modifying enzymes. SET-25 is a histone H3K9 methyltransferase (Towbin et al., 2012) that acts downstream of PRG-1 and has been implicated in the establishment of TEI (Ashe et al., 2012; Klosin et al., 2017). HPL-2, an H3K9me3 reader and homolog of mammalian HP1 (Couteau, 2002) is involved in transgene and piRNA-mediated gene silencing in the gonad (Ashe et al., 2012). Similar to prg-1 and the RdRP mutants, set-25 and hpl-2 mutants have normal P0 naïve attraction to and learned avoidance of PA14 but are specifically defective in F1 avoidance of PA14 (Fig. 5H, I). Together, these data suggest that prg-1/piRNAs and its downstream nuclear RNAi pathway function in the transgenerational inheritance of learned pathogen avoidance (Fig 5J, K).
Inheritance of learned avoidance of Pseudomonas confers survival advantages to progeny
Because we observed pathogenic avoidance behavior in wild-type animals rather than in mutants or an artificial situation, the transgenerational pathogenic avoidance paradigm may represent a natural context for such behaviors. Providing worms with the opportunity to avoid PA14 (by placing them on a small PA14 lawn, where there is some opportunity to avoid the pathogen, instead of a fully-covered plate) significantly increases their survival (Fig. 6A; 76%; p<0.0001). Perhaps more importantly, we find that naïve F1 progeny of PA14-trained mothers survive significantly longer (p=0.0013) on a small PA14 lawn compared to progeny of OP50-trained controls (Fig. 6B). The increased survival of progeny of trained mothers is dependent on the ability to avoid PA14, since obligate PA14 exposure (full PA14 lawn) confers no survival difference (Fig. 6C). Finally, prg-1 mutant F1s do not benefit from P0 training on PA14 (Fig. 6D), even when presented with the opportunity to avoid Pseudomonas. Thus, the PRG-1-dependent transgenerational inheritance of learned pathogenic avoidance is required for this enhanced survival.
Fig. 6: Inheritance of learned avoidance of Pseudomonas confers survival advantages to progeny.
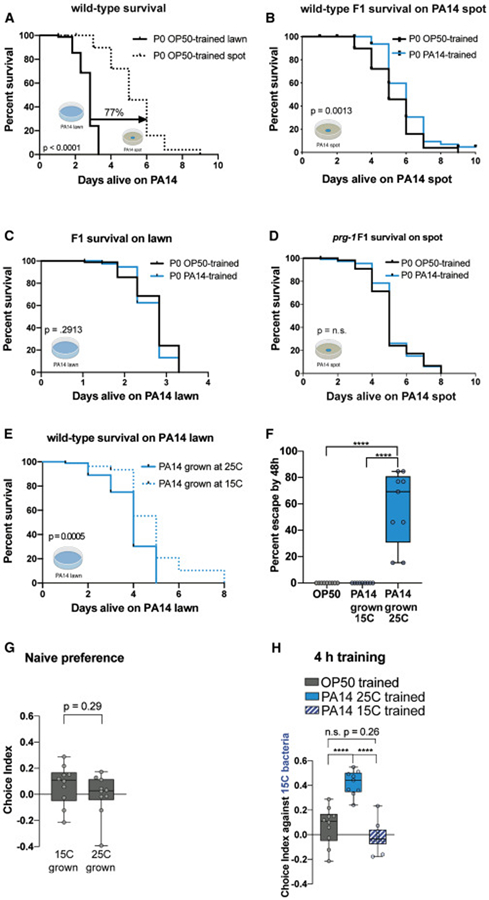
(A) Survival was measured on PA14 spots vs lawns. The ability to escape PA14 provides a survival advantage, compared to animals on a lawn of PA14 (p < 0.0001, 77% increase). (B) Progeny of PA14-trained mothers have a survival advantage on an escapable spot of PA14, compared to progeny of OP50-trained mothers (p = 0.0013). (C) No difference in survival of OP50- or PA14-trained mothers was observed when worms were unable to escape PA14. (D) prg-1 mutants do not exhibit the survival benefit on escapable lawns, unlike wild type (B). (E) PA14 grown at 15°C is less pathogenic than PA14 grown at 25°C. Bacteria was grown on plates at the indicated temperatures, and the survival assay was performed for all conditions at 20°C. (F) At 20°C, C. elegans leave pathogenic (25°C grown) PA14 at greater rates than from OP50 or less pathogenic (15°C grown) PA14. (G) Untrained (naïve) animals are attracted to PA14 regardless of the pathogenicity of PA14 (15°C grown vs 25°C grown). (H) 4h of training on virulent PA14 (25°C grown) is sufficient to cause maternal avoidance of less virulent (15°C grown). However, 4h of training on less virulent PA14 is not sufficient to elicit avoidance. All survival assays (A-E): Log-rank (Mantel-Cox) test. n = 80 worms. One-Way ANOVA, Tukey’s multiple comparison test, mean ± SEM. n ≥ 7 choice assay plates with 50-200 worms. ****p < 0.0001, ns = not significant.
PA14 exposure provides both parents and several generations of progeny with the ability to avoid this pathogen later, promoting survival. Why, then, are these advantageous behavioral responses not hard-wired into C. elegans’ naïve behavioral repertoire? Could it in fact be beneficial for the progeny of PA14-exposed parents to reacquire PA14 attraction later? To explore the hypothesis that loss of learned avoidance could be beneficial, we measured behavioral responses to PA14 grown under varying temperature conditions that alter bacterial virulence, as might be experienced in the wild. PA14 is pathogenic to worms when grown at elevated temperatures (25°C), but is less pathogenic when raised at lower temperatures (15°C) (Fig. 6E). Similarly, animals escape from a lawn of pathogenic PA14 (grown at 25°C), but not from less virulent PA14 (grown at 15°C), similar to their behavior on OP50 (Fig. 6F). Despite differences in the potential quality and pathogenicity of the food source, naïve worms similarly prefer PA14 raised at 25°C and 15°C compared to OP50 (Fig. 6G). Strikingly, while worms trained on 15°C-grown non-pathogenic bacteria do not avoid PA14, underscoring the importance of virulence in inducing avoidance, worms trained on pathogenic (25°C-grown) PA14 avoid the less-pathogenic (15°C-grown) PA14 (Fig. 6H). This result suggests that pathogenic Pseudomonas training causes worms to avoid all Pseudomonas, including non-pathogenic bacteria that may provide adequate nutrition. Therefore, “forgetting” the learned avoidance after a few generations may be necessary to allow the worms to once again be attracted to nutritious, non-pathogenic Pseudomonas.
Discussion
Animals exposed to pathogens employ several strategies to maintain fitness in the face of imminent death. C. elegans exposed to PA14 immediately engage innate immune responses (Troemel et al., 2006), and subsequently induce behavioral avoidance (Zhang et al., 2005). In fact, the ability to escape PA14 after direct contact promotes survival (Fig. 6A). Transgenerational epigenetic inheritance of learned PA14 avoidance may serve to prepare progeny and generations of grandprogeny for likely environmental PA14 exposure, so that they, too, can escape the pathogen. Our findings suggest that exposure to the pathogen PA14 engages a piRNA-dependent transgenerational inheritance network that confers multigenerational behavioral and gene expression changes; these changes may provide an adaptive survival advantage to progeny. The PA14-induced TEI behavior is dependent on PA14 virulence, lasts through the F4 generation, can be transmitted by either the male or female germline, and is distinct from previously-described pathogenic learning behaviors in C. elegans. We also found that the learned TEI behavior correlates with elevated daf-7p::gfp expression in the ASI neuron on the same generational time scale as the behavior, and that the learned ability to avoid PA14 results in a survival advantage in the progeny of trained parents—all phenotypes that are dependent on PIWI/PRG-1 and its downstream molecular components to execute transgenerational epigenetic inheritance.
By analyzing mutants for their roles in naïve P0 attraction to PA14, P0 learned avoidance of PA14 after 24h exposure, and the transgenerational inheritance of learned avoidance in the F1 progeny, we identified the molecular processes required for each step (Fig. 5J, K). In contrast to its previously identified role in L1 imprinting in the P0 generation (Jin et al., 2016), CREB is not required for any of the steps (Fig. 1F), likely because our testing of behavior occurs immediately after training in the P0s, rather than a long-term, spaced-training induced memory. Components of the COMPASS complex (SET-2, WDR-5.1, and RBR-2) are required for naïve preference for PA14, whereas components of the nuclear RNAi pathway (SET-32 and HRDE-1) are required for P0 pathogenic learning. Despite the fact that NRDE-4 can act with HRDE-1, NRDE-4’s requirement for normal naïve preference suggests that the NRDE-2/−4-independent HRDE-1 pathway may regulate pathogenic learning (Fig 4E). Finally, the transgenerational inheritance of pathogenic learning requires the activity of PRG-1/Piwi Argonaute, MUT-7/RNAse D, and RRF-1/RdRP (Fig. S6), which have been previously shown to work together to regulate 22G siRNA amplification (Phillips et al., 2012). These 22G siRNAs then guide the deposition and maintenance of histone marks through the activity of SET-25/H3K9 histone methyltransferase (Towbin et al., 2012) and the HPL-2/HP1 H3K9me3 reader (Couteau, 2002), which are also required for TEI of pathogen avoidance (Fig. 5J, K; Fig. S6).
Piwi/PRG-1-dependent piRNA pathways are potent mediators of TEI-related phenomena across a variety of metazoan species. In the germline of flies, worms, and mice, piRNAs regulate the stable silencing of germline transposons and are indispensable for germline development (Ghildiyal and Zamore, 2009). While piRNA biogenesis and secondary amplification mechanisms vary, the output of these Piwi-piRNA pathways converges on germline silencing through gene expression regulation (Ghildiyal and Zamore, 2009). Our results support the emerging model that in addition to the conserved, long-lasted silencing mediated by piRNAs (Ashe et al., 2012; Bagijn et al., 2012; Luteijn et al., 2012; Shirayama et al., 2012), this class of small RNAs is also involved in regulating transient transgenerational phenotypes (Brennecke et al., 2008; Grentzinger et al., 2012; Ashe et al., 2012), including pathogen avoidance behavior, as we have shown here.
Our results support a role for both sperm and oocytes in the transmission of learned pathogenic avoidance. There is substantial evidence supporting the role of mammalian sperm in TEI (Donkin and Barrès, 2018), with less direct evidence that oocytes can transmit transgenerational information; this is due to the difficulties of separating transgenerational mechanisms from environmental (i.e., in utero) influences in females, rather than the relative contributions of sperm and oocytes to TEI phenotypes. C. elegans hermaphrodites have higher nutritional needs and must make critical food choice decisions to support not only themselves, but also the development of their progeny. Furthermore, males of androdioecious species, such as C. elegans, are rare in nature (Ward and Carrel, 1979), and males prioritize exploration over food (Ryan et al., 2014). If hermaphrodites depended exclusively on male sperm for TEI of pathogenic learning, they might have no way of preparing progeny for potential PA14 exposure. By contrast, males that encounter PA14 and subsequently mate with naïve hermaphrodites may prepare progeny for potentially hazardous local PA14 environments. In effect, the ability of both parents to epigenetically warn progeny of the presence of pathogenic Pseudomonas could be an ideal survival strategy.
If PA14 avoidance promotes C. elegans fitness, why is this behavior not hard-wired, and what is the advantage of losing the TEI avoidance behavior after several generations? The Pseudomonas genus, which includes several non-pathogenic Pseudomonas species, makes up a large fraction - perhaps a third - of C. elegans’ natural environment (Samuel et al., 2016). While some Pseudomonas are detrimental to fitness, consumption of other Pseudomonas species increases C. elegans progeny production (Dirksen et al., 2016), suggesting that several Pseudomonae may provide a substantial source of nutrition. This appears true even among the same species of Pseudomonas, in that PA14 can exhibit variable pathogenicity depending on environmental conditions, such as temperature, and that virulent PA14-exposed worms cannot distinguish between harmful and potentially nutritious PA14 (Fig. 6E-H). Thus, parental and learned avoidance behavior in subsequent generations may only be beneficial to worms that may continue to encounter PA14 in their local environment. Therefore, naïve avoidance and indefinite transgenerational inheritance of avoidance of all Pseudomonas species might be a poor long-term strategy that could deprive animals of adequate nutrition. Instead, temporary avoidance of pathogenic Pseudomonas might drive the worms’ progeny to escape pathogenic food sources, while eventually allowing the return to consumption of potentially nutritious Pseudomonae. While germline silencing of transposons can last indefinitely, transgenerational epigenetic inheritance of transient phenotypes occurs over shorter generational timescales, typically lasting 1-4 generations in flies, worms, and mice (Perez and Lehner, 2019). While our work does not define the molecular events that govern how long a TEI phenotype lasts, it does provide biological insight into why both the acquisition and loss of a behavior several generations later would be beneficial. Together, our results demonstrate that transgenerational avoidance of pathogenic bacteria provides a biological context for TEI in C. elegans, where animals must distinguish between both beneficial and detrimental food sources to ensure survival of self and progeny in a dynamic environment.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Coleen T. Murphy (ctmurphy@princeton.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C. elegans and bacterial strains and cultivation
Strains were provided by the CGC. SX922: prg-1(n4357), RB1025: set-2(ok952), ZR1: rbr-2(tm1231), VC967: set-32(ok1457), FK181: ksIs2 [Pdaf-7::GFP + rol-6(su1006)], MT9973: crh-1(n3315), PY7505: oyIs84 [gpa-4p::TU#813 + gcy-27p::TU#814 + gcy-27p::GFP + unc-122p::DsRed], LC108: uIs69[(pCFJ90) myo-2p::mCherry + unc-119p::sid-1], NL1820: mut-7(pk720), MAH23: rrf-1(p1417), MT17463: set-25(n5021), RB995: hpl-2(ok916), YY453: nrde-4(gg129), CB4108: fog-2(q71). Strains were provided by the National Bioresource hrde-1(tm1200). Strain CQ605 (prg-1(n4357); ksIs2 [Pdaf-7::GFP + rol-6(su1006)] was made by mating SX922 with FK181. OP50 and OP50-1 were provided by the CGC. S. marcescens (ATCC 274) was purchased from ATCC. PA14 and LasR (Siryaporn et al., 2015) were gifts from Z. Gitai. Worm strains were maintained at 15°C on High Growth Media (HG) plates (3 g/L NaCl, 20 g/L Bacto-peptone, 30 g/L Bacto-agar in distilled water, with 4 mL/L cholesterol (5 mg/mL in ethanol), 1 mL/L 1M CaCl2, 1 mL/L 1M MgSO4, and 25 mL/L 1M potassium phosphate buffer (pH 6.0) added to molten agar after autoclaving) on E. coli OP50 using standard methods.
METHOD DETAILS
Pathogen training
Eggs from young adult hermaphrodites were obtained by bleaching (alkaline-bleach solution (e.g., 5.5 mL water, 1.5 mL 5N KOH, 3.0 mL sodium hypochlorite), followed by repeated washing of collected eggs in M9 buffer (6 g/L Na2HPO4, 3 g/L KH2PO4, 5 g/L NaCl and 1 mL/L 1M MgSO4 in distilled water), and were placed on to High Growth (HG) plates, and raised at 20°C for 2 days. Training plates were prepared by inoculating overnight cultures of OP50, PA14, or S. marcescens in LB at 37°C. Overnight cultures were diluted in LB to an Optical Density (OD600) = 1 and used to seed Nematode Growth Media (NGM) (NGM: 3 g/L NaCl, 2.5 g/L Bacto-peptone, 17 g/L Bacto-agar in distilled water, with 1 mL/L cholesterol (5 mg/mL in ethanol), 1 mL/L 1M CaCl2, 1 mL/L 1M MgSO4,and 25 mL/L 1M potassium phosphate buffer (pH 6.0) added to molten agar after autoclaving) plates. Plates were incubated at 25°C in separate incubators for 2 days. For 15°C experiments, PA14 was prepared by centrifuging 5 mL overnight cultures for 10 minutes at 5000 rpm. The supernatant was removed, and the remaining pellet was resuspended in 5 mL of fresh LB. Washed bacteria were used to inoculate (1:500) fresh LB to grow at 15°C for 2 days. Cultures were diluted in LB to an OD600 = 1 and used to seed NGM plates. Plates were incubated at 15°C for 2 days. On day of training (i.e., 2 days post bleaching), all training plates were left to cool on a bench top for < 1 hr. 10 μL of pooled L4 worms were plated onto OP50 seeded training plates, while 40 μL of worms were plated onto pathogen seeded training plates. Worms were incubated on training plates at 20°C in separate containers for 24h. After 24h, worms were washed off plates in M9 3x. Some worms were used for an aversive learning assay, while the majority of worms were bleached and eggs were placed onto HG plates at 20°C for 3 days. For experiments involving RNAi the standard HGM agar was supplemented with 1 mL/L 1M IPTG (isopropyl β-d-1-thiogalactopyranoside) and 1 mL/L 100 mg/mL carbenicillin; animals were trained using pL4440 empty vector as control RNAi in HT115 bacteria.
Aversive learning assay
Overnight bacterial cultures were diluted in LB to an Optical Density (OD600) = 1, and 25 μL of each bacterial suspension was seeded on a 60 mm NGM plate and incubated at 25°C for 2 days or 15°C for 2 days. After two days, assay plates were left at room temperature for 1 hr before use. Immediately before use, 1 μL of 1M sodium azide was spotted onto each bacterial lawn to be used as a paralyzing agent during choice assay. To start the assay (modified from Zhang et al., 2005), worms were washed off training plates in M9, and washed 2 additionally times in M9. 5 μL of worms were spotted at the bottom of the assay plate, using a wide orifice tip, midway between the bacterial lawns. Assays were incubated at room temperature for 1 hr before counting the number of worms on each lawn.
In experiments in which F1 and subsequent generations are used: All animals tested are washed off HG plates with M9 at Day 1. Some of the pooled animals are subjected to an aversive learning assay, while the majority of worms are bleached onto HG plates left at 20°C for 3 days and used to test F2s.
Odor + 2-aminoacetophenone (2AA) training
3 mL of fresh overnight bacterial cultures (OD600 OP50 = 1.4-1.7, OD600 PA14 = 3.3-3.5), water, or 2AA (catalog no. A38207, Sigma-Alderich) (1 mM, diluted in water) were placed into 2 lids of a 35 mm petri dish, respectively, which was then placed in the lid of an inverted 10 cm NGM petri dish that had been prepared as described for pathogen training. Odor training assays were left in the dark at room temperature for 24h. All training conditions were maintained in separate containers.
L1 imprinting
Eggs from young hermaphrodites were obtained by bleaching and placed directly onto OP50 or pathogen prepared training plates (5 μL of eggs were placed onto OP50 training plates, and 20 μL of eggs were placed onto PA14 training plates). Plates were incubated at 20°C for 24h. After 24h, worms were washed off training plates using M9 + 50 mg/mL streptomycin. Worms were washed 2 times and plated onto HGM+300 mg/mL streptomycin plates seeded with OP50-1 (streptomycin resistant OP50). Worms were left to mature to Day 1 adults and used in an aversive learning assay. Subsequent generations were prepared by bleaching pooled animals onto HG plates.
PA14 survival assay
OP50 and PA14 were grown in liquid culture and diluted as described above. For full lawn assays. 750 μL of diluted OP50 or PA14 was spread to completely cover a 10 cm NGM plate. For PA14 spot assays, 100 μL of diluted PA14 was placed in the center of a 10 cm NGM plate, resulting in a 2 cm spot in the center of the plate. Plates were incubated for 2 days at 25°C or 15°C to allow bacterial growth. Plates were equilibrated to 20°C before the addition of Day 1 worms to plates. Assays were performed at 20°C. PA14-lawn assays were counted every 6-8 h. PA14-spot assays were counted every 24h. Every 48h, worms in both assays were moved onto new plates. For spot assays, animals were transferred to similar locations on new plates.
fog-2 mating assay
L4 males or females were picked onto PA14 or OP50 training plates, prepared as described above. 24h after training, males and females were washed 2 times using M9 + 300 ug/mL streptomycin. Worms were then pipetted (in a ratio of 2:1 males to females) onto NGM + 300 ug/mL streptomycin plates, seeded the day before with 300 μL of OP50-1 (streptomycin resistant) bacteria in the following combinations: OP50 males x OP50 females, OP50 males x PA14 females, PA14 males x PA14 females, PA14 males x OP50 females. Worms were left to mate for 24h at 20°C. Following mating, worms were bleached and eggs were transferred onto OP50-seeded HG plates. Early Day 1 animals were tested in an aversive learning assay as previously described. Females and males were counted separately.
RNA Interference
RNAi experiments were conducted using the standard feeding RNAi method. Bacterial clones expressing the control (empty vector, pL4440) construct and the dsRNA targeting C. elegans genes were obtained from the Ahringer RNAi library. All RNAi clones were sequenced prior to use. RNAi-induced knockdown was conducted by bleaching progeny of HT115 pL4440 or PA14 trained animals onto RNAi seeded plates.
Antibodies
The primary H3K4me3 antibody (8580), and the primary H3K9me3 antibody (8898) were obtained from Abcam. The secondary antibody (A-11008) was obtained from Invitrogen.
Whole-mount immunofluorescence
Worms were dissected on positively-charged glass slides in 8 μL of 0.5 mM levamisole in M9 to paralyze worms followed by freeze-cracking on dry ice for 20 minutes. The worms were fixed by washing slides in cold methanol for 10 minutes followed by 5 minutes in acetone. The slides were then washed three times in PBST (PBS pH 7.4, 0.5% Triton X-100, 1mM EDTA) for 10 minutes. Following application of antibody, worms were covered with a 20 x 20-mm piece of parafilm and then incubated overnight at room temperature in a humidified-dark chamber. Primary antibodies used were H3K4me3 (Abcam 8580) or H3K9me3 (Abcam 8898) at 1:200 dilution. The next day slides were washed three times in PBST for 10 minutes. Secondary antibodies were added at 1:500 dilution (A-11008) and incubated overnight as before. The next day slides were again washed in PBST. Then 8 μLof vectasheild with DAPI was added to each slide and the coverslip was sealed with fingernail polish. Slides were visualized by Z-stack multi-channel (DIC, GFP) every 1 mm at 60X magnification. Images taken after whole-mount immunofluorescence are unable to be quantified because each condition analyzed must be dissected and stained on different slides, and staining varies slide to slide.
Imaging and fluorescence quantitation
Z-stack multi-channel (DIC, GFP) of day 1 adult GFP transgenic worms were imaged every 1 mm at 60X magnification; Maximum Intensity Projections and 3D reconstructions of head neurons, or germlines were built with Nikon NIS-Elements. To quantify daf-7p::GFP levels, worms were prepared and treated as described for pathogen training. Worms were mounted on agar pads and immobilized using 1 mM levamisole. GFP was imaged at 60X magnification and quantified using NIS-Elements software. Average pixel intensity was measured in each worm by drawing a bezier outline of the neuron cell body for 2 ASI head neurons and/or 2 ASJ head neurons.
RNA isolation
Adult worms (P0) and eggs (F1) were collected in M9 and washed several times to remove excess bacteria. Worm pellets were crushed in liquid nitrogen and transferred to an appropriate volume of Trizol LS. Total RNA was extracted from Trizol using the mirVana miRNA isolation kit (ThermoFisher). mRNA libraries for directional RNA sequencing were prepared using the SMARTer Apollo System and were sequenced (180-nt single-end) on the Illumina HiSeq 2000 platform. RNA samples for small RNA-seq were treated with 5’polyphosphatase (Lucigen) and prepared using the SMARTer Apollo system with modifications for small RNA library preparation. Briefly, sample fragmentation and initial bead size selection steps were omitted, and small RNA-containing libraries were Blue Pippin size selected (21-26nt insert size) prior to 75-nt single-end sequencing.
RNA-seq data analysis
FASTQC was used to assess read quality scores. Universal adapter sequences were trimmed from small RNA library sequences using Cutadapt v1.6 (Martin, 2011). Reads were mapped to the C. elegans genome (UCSC Feb 2013, ce11/ws245) using STAR (Dobin et al., 2013). Count matrices were generated using htseq-counts (mode: union). DESeq2 was used for differential expression analysis. Genes at FDR < 0.5 were considered significantly differentially expressed. Regularized log transformations were applied to the data to generate heatmaps. Sequences are deposited at NCBI BioProject PRJNA509938.
Gene Ontology analysis
g:Profiler (Reimand et al., 2016) was used to identify significantly enriched gene ontology (GO) terms from upregulated or downregulated gene lists (DESeq2 genes FDR < 1%). Biological Process GO terms were plotted using REVIGO (Supek et al., 2011). The size of the circle in each GO term plot reflects the overall size of the GO term category such that more general GO terms appear larger, while more specific GO terms are smaller. The color of the circle represents the significance of the GO term in the data set.
QUANTIFICATION AND STATISTICAL ANALYSIS
Lifespan assays were assessed using Kaplan-Meier log rank tests. For the comparison of choice indices between two genotypes (i.e., crh-1(n3315) vs wild-type), one-way ANOVA with Tukey’s multiple comparison test were used. For the comparison of choice indices between two generations in which only one genotype was tested (i.e., wild-type only), unpaired t tests were performed. For the comparison of learning indices between generations (i.e., in wild-type only), one-way ANOVA with Tukey’s multiple comparison test were used. For quantification of neuron intensity one-way ANOVA with Tukey’s multiple comparison test were used. Experiments were repeated on separate days with separate populations, to confirm that results were reproducible. Prism 8 software was used for all statistical analyses; software and further statistical details used for RNA-seq analyses are described in the Methods Details section of the STAR methods. Additional, statistical details of experiments, including sample size (with n representing the number of chemotaxis assays performed for behavior, number of individual neurons imaged, and number of worm used in survival assays) can be found in the figure legends.
DATA AVAILABILITY
Raw RNA-seq datasets are publically available through NCBI BioProject PRJNA509938 (https://www.ncbi.nlm.nih.gov/bioproject)
Supplementary Material
Fig. S1: Transgenerational inheritance of pathogen avoidance requires direct contact with PA14, Related to Figure 1.
(A) PA14 odor is not sufficient to induce aversive pathogenic learning. (B) 2-aminoacetophenone (2AA) is not sufficient to induce aversive pathogenic learning. One-Way ANOVA, Tukey’s multiple comparison test, mean ± SEM. n > 6-10 choice assay plates with 50-200 worms. *p ≤ 0.05, **p ≤ 0.01, ns = not significant. At least 2 biological replicates were performed for all aversive learning assays. (C) PA14 is lethal at 20°C. p < 0.0001, Log-rank (Mantel-Cox) test. n ≥ 78-81 worms per condition. (D) Adult male worms naively avoid PA14. One-Way ANOVA, Tukey’s multiple comparison test, mean ± SEM. n ≥ 6-10 choice assay plates with 50-200 worms). *p ≤ 0.05, **p ≤ 0.01, ns = not significant.
Fig. S2: Analysis of differentially expressed genes from P0 animals treated with PA14 vs OP50, Related to Figure 1.
(A-C) mRNAs differentially regulated in P0 animals after PA14 treatment. (A) Volcano plot and (B,C) heatmaps of PA14-upregulated (5829, red) and downregulated (4967, green). (A) Volcano plot of mRNAs shown at FDR < 5% and log2 fold change > and < 1. (B, C) Heatmaps of normalized log expression counts are shown for genes with FDR < 1% and base mean expression > 500. PA14-downregulated (D) and upregulated (E) genes (FDR < 1%) were analyzed for enriched gene ontology (GO) terms using g:Profiler, and significant GO terms were plotted using REVIGO.
Fig. S3: Analysis of differentially expressed genes from F1 animals treated with PA14 vs OP50, Related to Figure 2.
(A) F1 eggs were collected from PA14- or OP50-treated mothers. PA14-upregulated (4805, red) and downregulated (3536, green) mRNAs shown at FDR < 5% and log2 fold change > and < 1. (B) Heatmap of differentially expressed genes from F1 animals from OP50- or PA14-treated mothers. Normalized log expression counts are shown for all genes with FDR < 1% and base mean expression > 500. (C) PA14-upregulated and (D) downregulated genes (FDR < 1%) were analyzed for enriched gene ontology (GO) terms using g:Profiler, and significant GO terms were plotted using REVIGO. (E) PA14-upregulated genes in P0 and F1 animals (FDR < 5%) were examined for adult neuron expression (Kaletsky et al., 2016).
Fig. S4: Mutants that do not learn avoidance behavior exhibit similar behavior in F1 and F2 following P0 training, Related to Figure 4.
(A) F1 progeny of PA14-trained set-2, rbr-2, and (B) set-32 are defective in pathogenic aversive learning. (C) F2 progeny of PA14-trained set-2, rbr-2, and (D) set-32 are defective in pathogenic aversive learning. (E) F1 progeny of PA14-trained hrde-1 are defective in pathogenic aversive learning. (F) F1 progeny of PA14-trained nrde-4 have high naïve preference and are defective in pathogenic aversive learning. (G) F2 progeny from PA14-trained prg-1 grandmothers are defective in pathogenic aversive learning. One-Way ANOVA, Tukey’s multiple comparison test, mean ± SEM. n ≤ 6-10 choice assay plates with 50-200 worms per plate. *p ≤ 0.05, **p ≤ 0.01, ns = not significant. At least 2 biological replicates were performed for all aversive learning assays. Global H3K4me3 (H) and H3K9me3 (I) levels remain unchanged after exposure to PA14.
Fig. S5: RNAi-treated progeny of PA14-trained mothers still exhibit aversive learning defects in the F1 and F2 generations, Related to Figure 4.
(A) 24h of PA14 training in wild-type animals results in aversive learning compared to 24h of training with E. coli HT115. F1 progeny of wild-type PA14-trained mothers treated with set-2(RNAi) (B), set-32(RNAi) (C), hrde-1(RNAi) (D) were defective in pathogenic learning. (E) F2 hrde-1(RNAi) of PA14-trained mothers are defective in pathogenic learning. One-Way ANOVA, Tukey’s multiple comparison test, mean ± SEM. n = 10 choice assay plates with 50-200 worms per plate. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p < 0.0001, ns = not significant. At least 2 biological replicates were performed for all aversive learning assays.
Fig. S6: Molecular model of known TEI components, Related to Figure 5.
Summary of genes implicated in naïve preference, learning, and TEI avoidance based upon previous descriptions of protein function.
Supplementary Table 1. mRNA-seq results from P0 N2 on PA14 vs OP50, Related to Figure 1.
DESeq2 results from libraries prepared from P0 adult animals treated with PA14 or OP50 for 24h as described in the methods. Genes upregulated or downregulated by PA14 (FDR < 5%) are highlighted. g:Profiler gene ontology analysis was performed on differentially upregulated or downregulated genes (FDR < 1%). The complete results, including GO terms, KEGG pathways, Reactome terms, and enriched transcription factor motifs are shown.
Supplementary Table 2. mRNA-seq results from F1 N2 on PA14 vs OP50, Related to Figure 2.
DESeq2 results from libraries prepared from F1 eggs of mothers treated with PA14 or OP50 for 24h as described in the methods. Genes upregulated or downregulated by PA14 treatment of the mothers (FDR < 5%) are highlighted. g:Profiler gene ontology analysis was performed on differentially upregulated or downregulated genes (FDR < 1%). The complete results, including GO terms, KEGG pathways, Reactome terms, and enriched transcription factor motifs are shown.
Supplementary Table 3. Small RNAseq results from P0 N2 on PA14 vs OP50, Related to Figure 1.
DESeq2 results from small RNA libraries prepared from P0 adult animals treated with PA14 or OP50 for 24h as described in the methods. Small RNAs upregulated or downregulated by PA14 (FDR < 5%) are highlighted.
Highlights:
C. elegans transmit learned avoidance of P. aeruginosa for four generations
ASI and the TGF-β ligand daf-7 are required for transgenerational PA14 avoidance
Piwi/PRG-1 is required for transgenerational inheritance of P. aeruginosa avoidance
Transgenerational avoidance of P. aeruginosa provides fitness benefits to offspring
Acknowledgments:
We thank the C. elegans Genetics Center for strains; the Genomics Core Facility at Princeton University, J. Wiggins, and J. Miller for RNA-seq library preparation and sequencing; L. Parsons for assistance with data analysis; V. Cota for gonad dissections; W. Keyes, J. Ashraf, and R. Clausen for assistance counting aversive learning assays; Z. Gitai for bacteria strains; and the Murphy lab for discussion. CTM is the Director of the Glenn Center for Aging Research at Princeton and an HHMI-Simons Faculty Scholar.
Funding: RSM was supported by T32GM007388 (NIGMS), and further support was provided by a DP1 Pioneer Award to CTM (NIGMS 1DP2OD004402-01), The Glenn Foundation for Medical Research (GMFR CNV1001899), and the HHMI Faculty Scholar Program (AWD1005048).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests: Authors declare no competing interests.
References:
- Aoki K, Moriguchi H, Yoshioka T, Okawa K, and Tabara H (2007). In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 26, 5007–5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick E-M, Mitchell J, Bagijn MP, Cording AC, Doebley A-L, Goldstein LD, Lehrbach NJ, Le Pen J, et al. (2012). piRNAs Can Trigger a Multigenerational Epigenetic Memory in the Germline of C. elegans. Cell 150, 88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagijn MP, Goldstein LD, Sapetschnig A, Weick E-M, Bouasker S, Lehrbach NJ, Simard MJ, and Miska EA (2012). Function, Targets, and Evolution of Caenorhabditis elegans piRNAs. Science 337, 574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belicard T, Jareosettasin P, and Sarkies P (2018). The piRNA pathway responds to environmental signals to establish intergenerational adaptation to stress. BMC Biol. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, and Hannon GJ (2008). An Epigenetic Role for Maternally Inherited piRNAs in Transposon Silencing. Science 322, 1387–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A, and Kennedy S (2012). A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489, 447–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart KB, Guang S, Buckley BA, Wong L, Bochner AF, and Kennedy S (2011). A Pre-mRNA–Associating Factor Links Endogenous siRNAs to Chromatin Regulation. PLoS Genet. 7, e1002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton NO, Burkhart KB, and Kennedy S (2011). Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. Proc. Natl. Acad. Sci 108, 19683–19688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton NO, Furuta T, Webster AK, Kaplan REW, Baugh LR, Arur S, and Horvitz HR (2017). Insulin-like signalling to the maternal germline controls progeny response to osmotic stress. Nat. Cell Biol 19, 252–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F (2002). A heterochromatin protein 1 homologue in Caenorhabditis elegans acts in germline and vulval development. EMBO Rep. 3, 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L-L, Gao J-X, Zou C-G, Ma Y-C, and Zhang K-Q (2015). mir-233 Modulates the Unfolded Protein Response in C. elegans during Pseudomonas aeruginosa Infection. PLoS Pathog. 11, e1004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daxinger L, and Whitelaw E (2012). Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat. Rev. Genet 13, 153–162. [DOI] [PubMed] [Google Scholar]
- Dias BG, and Ressler KJ (2014). Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat. Neurosci. 17, 89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirksen P, Marsh SA, Braker I, Heitland N, Wagner S, Nakad R, Mader S, Petersen C, Kowallik V, Rosenstiel P, et al. (2016). The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model. BMC Biol. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkin I, and Barrès R (2018). Sperm epigenetics and influence of environmental factors. Mol. Metab 14, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, and Mello CC (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Gapp K, Jawaid A, Sarkies P, Bohacek J, Pelczar P, Prados J, Farinelli L, Miska E, and Mansuy IM (2014). Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat. Neurosci 17, 667–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, and Zamore PD (2009). Small silencing RNAs: an expanding universe. Nat. Rev. Genet 10, 94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Ucar D, Hauswirth AG, Mancini E, Lim JP, Benayoun BA, Shi Y, and Brunet A (2011). Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature 479, 365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer ER, Pérez CL, Van Gilst MR, Lee BH, and Ashrafi K (2008). Neural and Molecular Dissection of a C. elegans Sensory Circuit that Regulates Fat and Feeding. Cell Metab. 8, 118–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grentzinger T, Armenise C, Brun C, Mugat B, Serrano V, Pelisson A, and Chambeyron S (2012). piRNA-mediated transgenerational inheritance of an acquired trait. Genome Res. 22, 1877–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, and Bartel DP (2008). Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature 455, 1193–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A (2005). Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes Dev. 19, 683–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu SG, Pak J, Guang S, Maniar JM, Kennedy S, and Fire A (2012). Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat. Genet 44, 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, and Kennedy S (2008). An Argonaute Transports siRNAs from the Cytoplasm to the Nucleus. Science 321, 537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha H, Hendricks M, Shen Y, Gabel CV, Fang-Yen C, Qin Y, Colón-Ramos D, Shen K, Samuel ADT, and Zhang Y (2010). Functional Organization of a Neural Network for Aversive Olfactory Learning in Caenorhabditis elegans. Neuron 68, 1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heestand B, Simon M, Frenk S, Titov D, and Ahmed S (2018). Transgenerational Sterility of Piwi Mutants Represents a Dynamic Form of Adult Reproductive Diapause. Cell Rep. 23, 156–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbert ZA, and Kim DH (2017). Sexually dimorphic control of gene expression in sensory neurons regulates decision-making behavior in C. elegans. ELife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Pokala N, and Bargmann CI (2016). Distinct Circuits for the Formation and Retrieval of an Imprinted Olfactory Memory. Cell 164, 632–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletsky R, Lakhina V, Arey R, Williams A, Landis J, Ashraf J, and Murphy CT (2016). The C. elegans adult neuronal IIS/FOXO transcriptome reveals adult phenotype regulators. Nature 529, 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman AL, Ashraf JM, Corces-Zimmerman MR, Landis JN, and Murphy CT (2010). Insulin Signaling and Dietary Restriction Differentially Influence the Decline of Learning and Memory with Age. PLoS Biol. 8, e1000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF, Haverkamp TH, van Luenen HGA., and Plasterk RH (1999). mut-7 of C. elegans, Required for Transposon Silencing and RNA Interference, Is a Homolog of Werner Syndrome Helicase and RNaseD. Cell 99, 133–141. [DOI] [PubMed] [Google Scholar]
- Kishimoto S, Uno M, Okabe E, Nono M, and Nishida E (2017). Environmental stresses induce transgenerationally inheritable survival advantages via germline-to-soma communication in Caenorhabditis elegans. Nat. Commun 8, 14031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosin A, Casas E, Hidalgo-Carcedo C, Vavouri T, and Lehner B (2017). Transgenerational transmission of environmental information in C. elegans. Science 356, 320–323. [DOI] [PubMed] [Google Scholar]
- Kudlow BA, Zhang L, and Han M (2012). Systematic Analysis of Tissue-Restricted miRISCs Reveals a Broad Role for MicroRNAs in Suppressing Basal Activity of the C. elegans Pathogen Response. Mol. Cell 46, 530–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, and Mylonakis E (2017). An Intestine-Derived Neuropeptide Controls Avoidance Behavior in Caenorhabditis elegans. Cell Rep. 20, 2501–2512. [DOI] [PubMed] [Google Scholar]
- Liu F, He C-X, Luo L-J, Zou Q-L, Zhao Y-X, Saini R, Han S-F, Knolker H-J, Wang L-S, and Ge B-X (2013). Nuclear Hormone Receptor Regulation of MicroRNAs Controls Innate Immune Responses in C. elegans. PLoS Pathog. 9, e1003545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn MJ, van Bergeijk P, Kaaij LJT, Almeida MV, Roovers EF, Berezikov E, and Ketting RF (2012). Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans: Extremely stable Piwi-induced gene silencing. EMBO J. 31, 3422–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Niu R, Huang T, Shao L-W, Peng Y, Ding W, Wang Y, Jia G, He C, Li C-Y, et al. (2018). N6-methyldeoxyadenine is a transgenerational epigenetic signal for mitochondrial stress adaptation. Nat. Cell Biol [DOI] [PubMed] [Google Scholar]
- Ma Y-C, Zhang L, Dai L-L, Khan RU, and Zou C-G (2017). mir-67 regulates P. aeruginosa avoidance behavior in C. elegans. Biochem. Biophys. Res. Commun 494, 120–125. [DOI] [PubMed] [Google Scholar]
- Mao H, Zhu C, Zong D, Weng C, Yang X, Huang H, Liu D, Feng X, and Guang S (2015). The Nrde Pathway Mediates Small-RNA-Directed Histone H3 Lysine 27 Trimethylation in Caenorhabditis elegans. Curr. Biol 25, 2398–2403. [DOI] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.Journal 17, 10. [Google Scholar]
- Meisel JD, Panda O, Mahanti P, Schroeder FC, and Kim DH (2014). Chemosensation of Bacterial Secondary Metabolites Modulates Neuroendocrine Signaling and Behavior of C. elegans. Cell 159, 267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi FK, and Prahlad V (2017). Olfactory experience primes the heat shock transcription factor HSF-1 to enhance the expression of molecular chaperones in C. elegans. Sci. Signal 10, eaan4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J, and Fire A (2007). Distinct Populations of Primary and Secondary Effectors During RNAi in C. elegans. Science 315, 241–244. [DOI] [PubMed] [Google Scholar]
- Pereira A, Gracida X, Kagias K, and Zhang Y (2019). Learning of pathogenic bacteria in adult C. elegans bidirectionally regulates pathogen response in the progeny. BioRxiv. [Google Scholar]
- Perez MF, and Lehner B (2019). Intergenerational and transgenerational epigenetic inheritance in animals. Nat. Cell Biol 21, 143–151. [DOI] [PubMed] [Google Scholar]
- Perez MF, Francesconi M, Hidalgo-Carcedo C, and Lehner B (2017). Maternal age generates phenotypic variation in Caenorhabditis elegans. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CM, Montgomery TA, Breen PC, and Ruvkun G (2012). MUT-16 promotes formation of perinuclear Mutator foci required for RNA silencing in the C. elegans germline. Genes Dev. 26, 1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol N, Link EM, Liu LX, Kurz CL, Alloing G, Tan M-W, Ray KP, Solari R, Johnson CD, and Ewbank JJ (2001). A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr. Biol 11, 809–821. [DOI] [PubMed] [Google Scholar]
- Rechavi O, Minevich G, and Hobert O (2011). Transgenerational Inheritance of an Acquired Small RNA-Based Antiviral Response in C. elegans. Cell 147, 1248–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechavi O, Houri-Ze’evi L, Anava S, Goh WSS, Kerk SY, Hannon GJ, and Hobert O (2014). Starvation-Induced Transgenerational Inheritance of Small RNAs in C. elegans. Cell 158, 277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J, Arak T, Adler P, Kolberg L, Reisberg S, Peterson H, and Vilo J (2016). g:Profiler—a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 44, W83–W89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z, and Ambros VR (2015). Caenorhabditis elegans microRNAs of the let-7 family act in innate immune response circuits and confer robust developmental timing against pathogen stress. Proc. Natl. Acad. Sci. U. S. A 112, E2366–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan DA, Miller RM, Lee K, Neal SJ, Fagan KA, Sengupta P, and Portman DS (2014). Sex, Age, and Hunger Regulate Behavioral Prioritization through Dynamic Modulation of Chemoreceptor Expression. Curr. Biol 24, 2509–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel BS, Rowedder H, Braendle C, Félix M-A, and Ruvkun G (2016). Caenorhabditis elegans responses to bacteria from its natural habitats. Proc. Natl. Acad. Sci 113, E3941–E3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedl T, and Kimble J (1988). fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics 119, 43–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulenburg H, and Félix M-A (2017). The Natural Biotic Environment of Caenorhabditis elegans. Genetics 206, 55–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee H-C, Gu W, Ishidate T, Conte D, and Mello CC (2012). piRNAs Initiate an Epigenetic Memory of Nonself RNA in the C. elegans Germline. Cell 150, 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtonda BB, and Avery L (2006). Dietary choice behavior in Caenorhabditis elegans. J. Exp. Biol 209, 89–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RHA, and Fire A (2001). On the Role of RNA Amplification in dsRNA-Triggered Gene Silencing. Cell 107, 465–476. [DOI] [PubMed] [Google Scholar]
- Sijen T, Steiner FA, Thijssen KL, and Plasterk RHA (2007). Secondary siRNAs Result from Unprimed RNA Synthesis and Form a Distinct Class. Science 315, 244–247. [DOI] [PubMed] [Google Scholar]
- Simon M, Sarkies P, Ikegami K, Doebley A-L, Goldstein LD, Mitchell J, Sakaguchi A, Miska EA, and Ahmed S (2014). Reduced Insulin/IGF-1 Signaling Restores Germ Cell Immortality to Caenorhabditis elegans Piwi Mutants. Cell Rep. 7, 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siryaporn A, Kim MK, Shen Y, Stone HA, and Gitai Z (2015). Colonization, Competition, and Dispersal of Pathogens in Fluid Flow Networks. Curr. Biol 25, 1201–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, and Šmuc T (2011). REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 6, e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MW, Mahajan-Miklos S, and Ausubel FM (1999). Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. U. S. A 96, 715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauffenberger A, and Parker JA (2014). Heritable Transmission of Stress Resistance by High Dietary Glucose in Caenorhabditis elegans. PLoS Genet. 10, e1004346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin BD, González-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, and Gasser SM (2012). Step-Wise Methylation of Histone H3K9 Positions Heterochromatin at the Nuclear Periphery. Cell 150, 934–947. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, and Kim DH (2006). p38 MAPK Regulates Expression of Immune Response Genes and Contributes to Longevity in C. elegans. PLoS Genet. 2, e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassoler FM, White SL, Schmidt HD, Sadri-Vakili G, and Pierce RC (2013). Epigenetic inheritance of a cocaine-resistance phenotype. Nat. Neurosci 16, 42–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Brunschwig K, Okihara KL, Muller F, Tijsterman M, and Plasterk RHA (2006). Long-term gene silencing by RNAi: Gene expression. Nature 442, 882–882. [DOI] [PubMed] [Google Scholar]
- Ward S, and Carrel JS (1979). Fertilization and sperm competition in the nematodeCaenorhabditis elegans. Dev. Biol 73, 304–321. [DOI] [PubMed] [Google Scholar]
- Wianny F, and Zernicka-Goetz M (2000). Specific interference with gene function by double-stranded RNA in early mouse development. Nat. Cell Biol 2, 70–75. [DOI] [PubMed] [Google Scholar]
- Woodhouse RM, Buchmann G, Hoe M, Harney D, Larance M, Boag PR, and Ashe A (2018a). The chromatin modifiers SET-25 and SET-32 are required for initiation but not long-term maintenance of transgenerational epigenetic inheritance. [DOI] [PubMed] [Google Scholar]
- Woodhouse RM, Buchmann G, Hoe M, Harney DJ, Low JKK, Larance M, Boag PR, and Ashe A (2018b). Chromatin Modifiers SET-25 and SET-32 Are Required for Establishment but Not Long-Term Maintenance of Transgenerational Epigenetic Inheritance. Cell Rep. 25, 2259–2272.e5. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lu H, and Bargmann CI (2005). Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature 438, 179–184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1: Transgenerational inheritance of pathogen avoidance requires direct contact with PA14, Related to Figure 1.
(A) PA14 odor is not sufficient to induce aversive pathogenic learning. (B) 2-aminoacetophenone (2AA) is not sufficient to induce aversive pathogenic learning. One-Way ANOVA, Tukey’s multiple comparison test, mean ± SEM. n > 6-10 choice assay plates with 50-200 worms. *p ≤ 0.05, **p ≤ 0.01, ns = not significant. At least 2 biological replicates were performed for all aversive learning assays. (C) PA14 is lethal at 20°C. p < 0.0001, Log-rank (Mantel-Cox) test. n ≥ 78-81 worms per condition. (D) Adult male worms naively avoid PA14. One-Way ANOVA, Tukey’s multiple comparison test, mean ± SEM. n ≥ 6-10 choice assay plates with 50-200 worms). *p ≤ 0.05, **p ≤ 0.01, ns = not significant.
Fig. S2: Analysis of differentially expressed genes from P0 animals treated with PA14 vs OP50, Related to Figure 1.
(A-C) mRNAs differentially regulated in P0 animals after PA14 treatment. (A) Volcano plot and (B,C) heatmaps of PA14-upregulated (5829, red) and downregulated (4967, green). (A) Volcano plot of mRNAs shown at FDR < 5% and log2 fold change > and < 1. (B, C) Heatmaps of normalized log expression counts are shown for genes with FDR < 1% and base mean expression > 500. PA14-downregulated (D) and upregulated (E) genes (FDR < 1%) were analyzed for enriched gene ontology (GO) terms using g:Profiler, and significant GO terms were plotted using REVIGO.
Fig. S3: Analysis of differentially expressed genes from F1 animals treated with PA14 vs OP50, Related to Figure 2.
(A) F1 eggs were collected from PA14- or OP50-treated mothers. PA14-upregulated (4805, red) and downregulated (3536, green) mRNAs shown at FDR < 5% and log2 fold change > and < 1. (B) Heatmap of differentially expressed genes from F1 animals from OP50- or PA14-treated mothers. Normalized log expression counts are shown for all genes with FDR < 1% and base mean expression > 500. (C) PA14-upregulated and (D) downregulated genes (FDR < 1%) were analyzed for enriched gene ontology (GO) terms using g:Profiler, and significant GO terms were plotted using REVIGO. (E) PA14-upregulated genes in P0 and F1 animals (FDR < 5%) were examined for adult neuron expression (Kaletsky et al., 2016).
Fig. S4: Mutants that do not learn avoidance behavior exhibit similar behavior in F1 and F2 following P0 training, Related to Figure 4.
(A) F1 progeny of PA14-trained set-2, rbr-2, and (B) set-32 are defective in pathogenic aversive learning. (C) F2 progeny of PA14-trained set-2, rbr-2, and (D) set-32 are defective in pathogenic aversive learning. (E) F1 progeny of PA14-trained hrde-1 are defective in pathogenic aversive learning. (F) F1 progeny of PA14-trained nrde-4 have high naïve preference and are defective in pathogenic aversive learning. (G) F2 progeny from PA14-trained prg-1 grandmothers are defective in pathogenic aversive learning. One-Way ANOVA, Tukey’s multiple comparison test, mean ± SEM. n ≤ 6-10 choice assay plates with 50-200 worms per plate. *p ≤ 0.05, **p ≤ 0.01, ns = not significant. At least 2 biological replicates were performed for all aversive learning assays. Global H3K4me3 (H) and H3K9me3 (I) levels remain unchanged after exposure to PA14.
Fig. S5: RNAi-treated progeny of PA14-trained mothers still exhibit aversive learning defects in the F1 and F2 generations, Related to Figure 4.
(A) 24h of PA14 training in wild-type animals results in aversive learning compared to 24h of training with E. coli HT115. F1 progeny of wild-type PA14-trained mothers treated with set-2(RNAi) (B), set-32(RNAi) (C), hrde-1(RNAi) (D) were defective in pathogenic learning. (E) F2 hrde-1(RNAi) of PA14-trained mothers are defective in pathogenic learning. One-Way ANOVA, Tukey’s multiple comparison test, mean ± SEM. n = 10 choice assay plates with 50-200 worms per plate. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p < 0.0001, ns = not significant. At least 2 biological replicates were performed for all aversive learning assays.
Fig. S6: Molecular model of known TEI components, Related to Figure 5.
Summary of genes implicated in naïve preference, learning, and TEI avoidance based upon previous descriptions of protein function.
Supplementary Table 1. mRNA-seq results from P0 N2 on PA14 vs OP50, Related to Figure 1.
DESeq2 results from libraries prepared from P0 adult animals treated with PA14 or OP50 for 24h as described in the methods. Genes upregulated or downregulated by PA14 (FDR < 5%) are highlighted. g:Profiler gene ontology analysis was performed on differentially upregulated or downregulated genes (FDR < 1%). The complete results, including GO terms, KEGG pathways, Reactome terms, and enriched transcription factor motifs are shown.
Supplementary Table 2. mRNA-seq results from F1 N2 on PA14 vs OP50, Related to Figure 2.
DESeq2 results from libraries prepared from F1 eggs of mothers treated with PA14 or OP50 for 24h as described in the methods. Genes upregulated or downregulated by PA14 treatment of the mothers (FDR < 5%) are highlighted. g:Profiler gene ontology analysis was performed on differentially upregulated or downregulated genes (FDR < 1%). The complete results, including GO terms, KEGG pathways, Reactome terms, and enriched transcription factor motifs are shown.
Supplementary Table 3. Small RNAseq results from P0 N2 on PA14 vs OP50, Related to Figure 1.
DESeq2 results from small RNA libraries prepared from P0 adult animals treated with PA14 or OP50 for 24h as described in the methods. Small RNAs upregulated or downregulated by PA14 (FDR < 5%) are highlighted.
Data Availability Statement
Raw RNA-seq datasets are publically available through NCBI BioProject PRJNA509938 (https://www.ncbi.nlm.nih.gov/bioproject)


