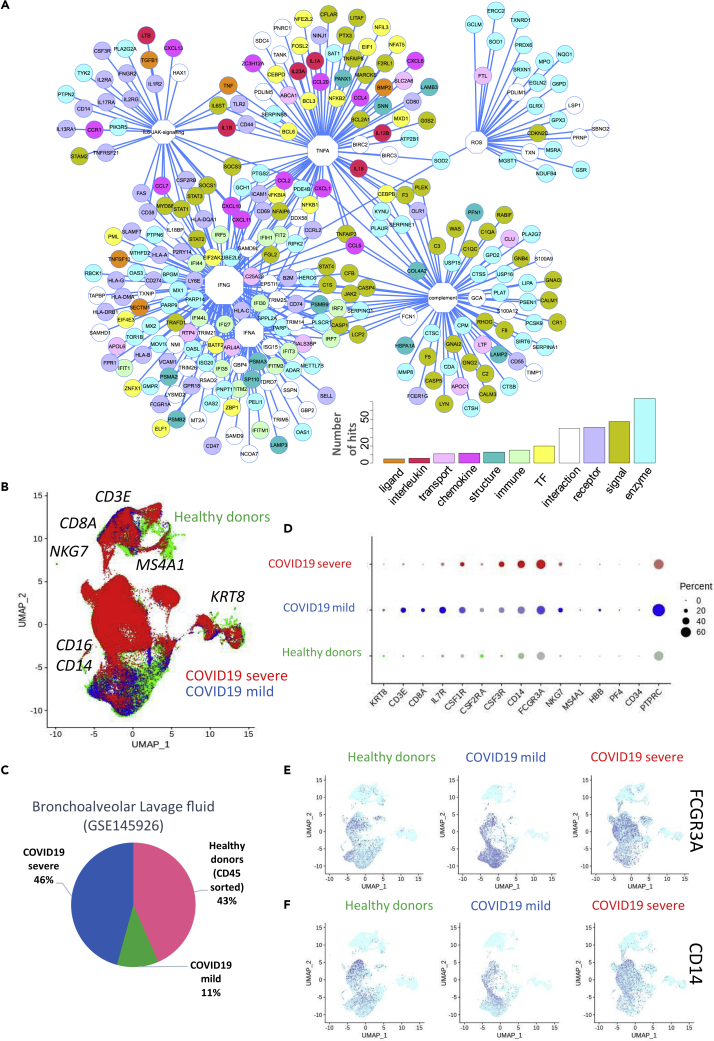
Figure 1.
Immune Network and Immune Checkpoint Regulation in COVID-19 Lung Samples
(A) Functional network of immune genes that were upregulated in lung samples from patients with COVID-19 compared with those from healthy donors; functions are represented as octagons, and their size represents the number of direct edge connections; (B) UMAP dimensionality reduction performed on merged single-cell transcriptome data from bronchoalveolar lavage fluid of healthy donors (green), patients with mild COVID-19 (blue), and patients with severe COVID-19 (red); major markers of cell subpopulations are indicated in black; (C) Pie chart representing the proportions of different cell types in the merged single-cell transcriptome analysis; (D) Dot plot of single-cell expression patterns (bronchoalveolar lavage fluid) of the main markers of each cluster, by patient of origin (HD, mild COVID-19, and severe COVID-19) (percent: percent of cells expressing each marker, expression level: color intensity), (E) Dimensionally reduced (UMAP) single-cell transcriptome (bronchoalveolar lavage fluid) expression patterns for the monocyte markers FCGR3A (CD16) and CD14 (F), by patient of origin (HD, mild COVID-19, and severe COVID-19).