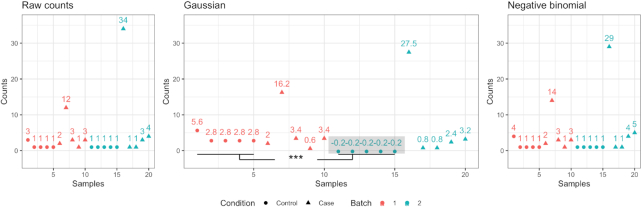
Figure 2.
Problematic results caused by applying a Gaussian-based batch adjustment on count data. We simulated a count matrix with a balanced case-control design and two batches. The first panel shows the counts for a simulated gene which is expressed at low levels in most cases and control samples. However, one case sample in each batch, especially in the second batch, contains a large value. Adjustment based on a Gaussian distribution brings the mean of the two batches to the same level, causing artificially induced differences across control samples from the two batches (P-value = 0.0033). When applying ComBat-seq based on negative binomial distribution, the adjusted data no longer contain the negative values (shown in gray box) or the erroneous significant difference between control samples from the two batches.