Fig. 2. MD simulation of DIII in solution.
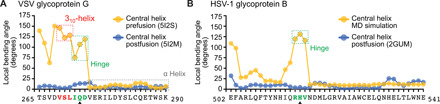
(A) Local bending angles of VSV-G DII in prefusion (yellow) (12) and postfusion (blue) (11) conformation. Protein Data Bank (PDB) numbers are given in parentheses. Amino acids are given in one-letter code. The 310-helix and hinge region are highlighted by dashed boxes in red and green, respectively. The α-helical part is highlighted by dashed box in gray. The position permitting a helix breaking mutation is marked with a black arrowhead. (B) Local bending angles of HSV-1 gB DIII in MD simulation (yellow) and postfusion (6) conformation (blue). The predicted hinge region in the MD simulation is marked in green. The position permitting a helix breaking mutation is marked with a black arrowhead.
