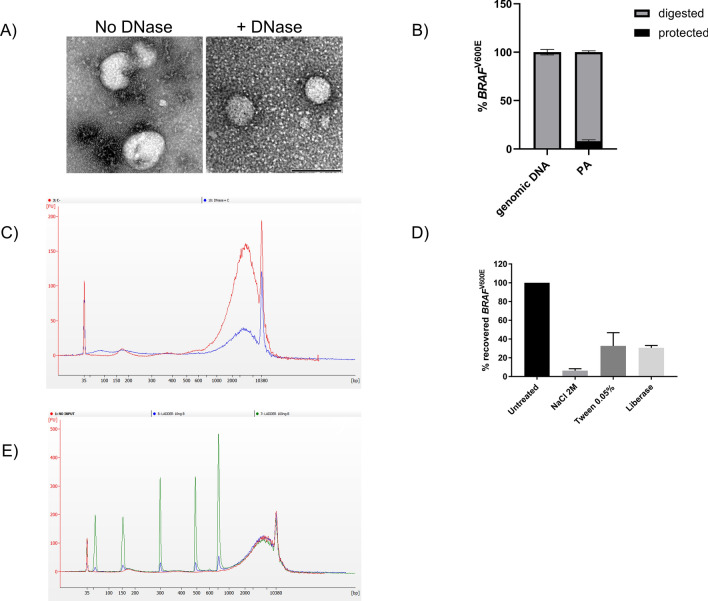
Figure 2.
PA efficiently captures both EV surface bound DNA and cell-free DNA. (A) Trasmission electron microscopy (TEM) analysis of EV pellets obtained with PA isolation prior and after Dnase I digestion. DNase-sensitive aggregates (negative staining) were observed in close proximity to the EV surface and in EV-free areas. Images are representative of three independent experiments. Size bar (black) is 100 nm. (B) Quantitation of protected mutant DNA in PA pellet. DNA was extracted and used to detect mutant BRAF by AS-QPCR. Genomic DNA from a commercial supplier was used as internal control. Data were expressed as percentage of digested or protected mutation and representative of three independent experiments. (C) Electropherogram analysis of DNA obtained before (red line) and after Dnase I digestion (blue line) of PA-derived pellet. Graph is representative of three independent experiments. (D) Recovery of BRAFV600E-positive extracellular vescles (EVs) from healthy donor plasma samples after peptide affinity (PA) isolation in the presence of NaCl (2 M), Tween-20 (0.05%) or enzymatic digestion with liberase. Following DNA extraction, mutant BRAF was detected by allele-specific quantitative PCR (AS-QPCR). Results are representative of three independent experiments. (E) Electropherogram analysis of 10 and 100 ng of ladder DNA spiked into healthy donor plasma (blue and green line respectively; no input sample red line) and isolated by PA. Fragment size were 50 bp, 150 bp, 300 bp, 500 bp, 766 bp. Electropherogram is representative of three independent experiments.