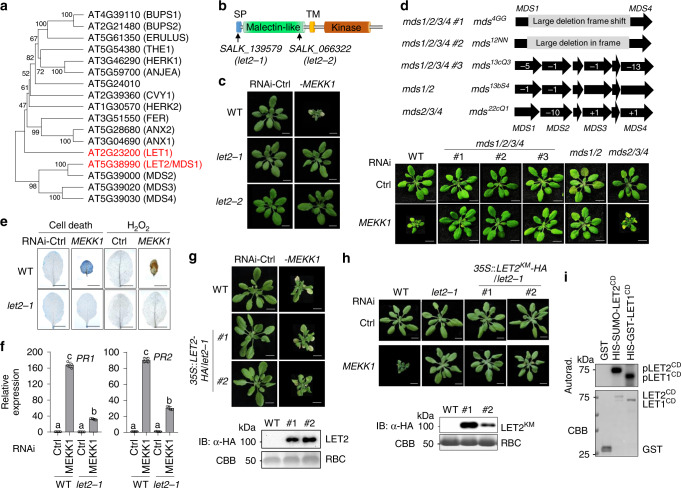
Fig. 1. LET2/MDS1 is involved in RNAi-MEKK1 cell death.
a A phylogenetic tree of CrRLK1L family proteins. The full-length protein sequences were used to generate the phylogenetic tree by the UPGMA method with 1000 bootstrap replicates in MEGA-X. b A schematics depicting LET2/MDS1 protein motifs and T-DNA insertion sites in the let2 mutants. LET2/MDS1 consists of the N-terminal signal peptide (SP), a malectin-like domain, a transmembrane domain (TM), and a cytosolic kinase domain. The arrows indicate the T-DNA insertion sites of the indicated mutant alleles. c The let2 mutants suppress plant dwarfism and leaf chlorosis induced by silencing MEKK1. The plant images were photographed at 3 weeks after inoculation with Agrobacterium carrying the indicated VIGS vectors. Ctrl is the vector containing GFP. Scale bar, 1 cm. d The LET2/MDS1, but not MDS2, MDS3, nor MDS4,is involved in RNAi-MEKK1 cell death. The mds1/2/3/4, mds1/2, and mds2/3/4 CRISPR/Cas knockout plants were inoculated with Agrobacterium carrying the indicated VIGS vectors. The plant images were photographed at 3 weeks after inoculation. The chromosome locations and Indels of MDS1, MDS2, MDS3, and MDS4 on individual mutants are shown on the top. Scale bar, 1 cm. e The let2-1 mutant suppresses cell death and H2O2 accumulation triggered by silencing MEKK1. The leaves were detached from plants in c and stained by trypan blue for cell death (left panel) and DAB for H2O2 accumulation (right panel). Scale bar, 0.5 cm. f The let2-1 mutant suppresses the expression of PR genes triggered by silencing MEKK1. The expression of PR1 and PR2 from plants in c was normalized to the expression of UBQ10 and the data are shown as the mean ± SE of four biological repeats (n = 4). P = 3.00 × 10−14 (PR1, column 1 and 2), P = 1.60 × 10−7 (PR1, column 3 and 4), P = 4.40 × 10−14 (PR1, column 2 and 4), P = 3.00 × 10−14 (PR2, column 1 and 2), P = 3.45 × 10−11 (PR2, column 3 and 4), and P = 5.10 × 10−14 (PR2, column 2 and 4). The different letters indicate the significant difference determined by one-way analysis of variance (ANOVA) followed by the Tukey test (P < 0.05). g Expression of LET2/MDS1 in let2-1 restores the cell death triggered by silencing MEKK1. The plant images were taken at 3 weeks after inoculation with Agrobacterium carrying the indicated VIGS vectors. #1 and #2 are two independent 35 S::LET2-HA transgenic lines in let2-1. Scale bar, 1 cm. Protein expression of LET2-HA in transgenic lines is shown on the bottom. The total proteins were immunoblotted by an α-HA antibody (upper panel). Coomassie Brilliant Blue (CBB) staining of RuBisCO (RBC) is shown as a loading control (lower panel). The molecular weight (MW) was labeled on the left of immunoblots as kDa. h The LET2/MDS1 kinase mutant cannot complement let2-1. Two lines of transgenic plants carrying the kinase-inactive mutant of LET2KM (K554E) driven by a 35S promoter in let2-1 are shown. Scale bar, 1 cm. Protein expression of LET2KM-HA in transgenic lines is shown on the bottom. i LET2/MDS1 bears kinase activity in vitro. GST and the LET2/MDS1 cytosolic kinase domain (HIS-SUMO-LET2CD) proteins were purified from E. coli. The LET1 cytosolic kinase domain (HIS-GST-LET1CD) proteins were purified from insect cells. The kinase assay was performed with [γ-32P] ATP. CBB staining was used as a loading control. The above experiments were repeated three times with similar results.