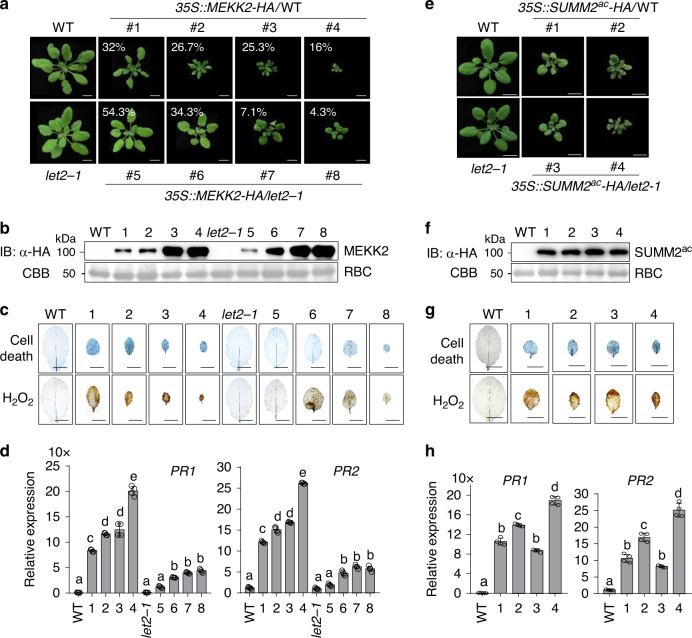
Fig. 3. LET2/MDS1 is required for MEKK2-, but not SUMM2ac-mediated autoimmunity.
a Plant dwarfisms and growth defects triggered by overexpressing MEKK2 in WT are alleviated in the let2-1 mutant. 75 and 70 independent primary (T1) transgenic plants carrying 35S::MEKK2-HA in WT and let2-1 were characterized, respectively. Four-week-old plants representing different levels of dwarfisms labeled with the percentage of the cognate category are shown. Scale bar, 1 cm. b Protein expression of MEKK2-HA in transgenic plants. Total proteins were isolated from plants in a and immunoblotted using an α-HA antibody (top panel). CBB staining for RBC is shown as the loading control (bottom panel). c The cell death and H2O2 accumulation triggered by overexpressing MEKK2 in WT are reduced in the let2-1 mutant. Leaves from plants in a were stained by trypan blue for cell death (upper panel) and DAB for H2O2 (lower panel). Scale bar, 0.5 cm. d The elevated expression of PR1 and PR2 triggered by overexpressing MEKK2 in WT is reduced in let2-1. The expression of PR1 and PR2 was normalized to the expression of UBQ10 and the data are shown as the mean ± SE of four biological repeats (n = 4). The different letters indicate the significant difference determined by one-way ANOVA followed by the Tukey test (P < 0.05). The plants 1–4 are 35S::MEKK2-HA/WT, and 5–8 are 35S::MEKK2-HA/let2-1 (a–d). e Plant dwarfisms and growth defects triggered by overexpressing SUMM2ac are similar in WT and let2-1. In all, 68 and 71 independent primary (T1) transgenic plants carrying 35S::SUMM2ac-HA in WT and let2-1 were characterized respectively. Two represenstative 3-week-old plants, which showed the growth defects, and their controls, are shown in the figure. Scale bar, 1 cm. f Protein expression of SUMM2ac-HA in transgenic plants. g The cell death and H2O2 accumulation triggered by overexpressing SUMM2ac in WT and let2-1. h The expression levels of PR1 and PR2 triggered by overexpressing SUMM2ac in WT and let2-1. The expression of PR1 and PR2 was normalized to the expression of UBQ10 and the data are shown as the mean ± SE of four biological repeats (n = 4). P < 1.00 × 10−15 (PR1, column 1 and 2), P < 1.00 × 10−15 (PR1, column 1 and 3), P = 2.62 × 10−12 (PR1, column 1 and 4), P < 1.00 × 10−15 (PR1, column 1 and 5), P = 1.24 × 10−7 (PR2, column 1 and 2), P = 1.04 × 10−10 (PR2, column 1 and 3), P = 5.86 × 10−6 (PR2, column 1 and 4), P < 1.00 × 10−15 (PR2, column 1 and 5). The different letters indicate the significant difference determined by one-way ANOVA followed by the Tukey test (P < 0.05). The plants 1–2 are 35S::SUMM2ac-HA/WT, and 3–4 are 35S::SUMM2ac-HA/let2-1 (e–h). The above experiments were repeated three times with similar results.