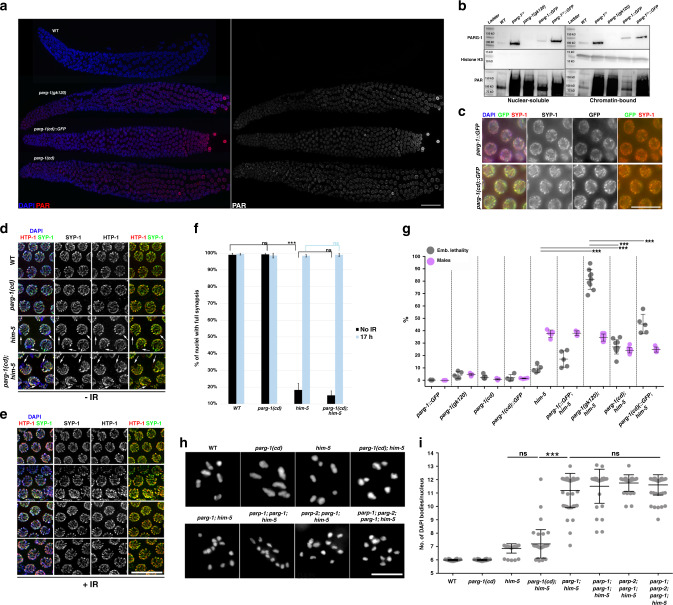
Fig. 6. PARG-1 catalytic activity is dispensable for recombination.
a Left: whole-mount germlines of the indicated genotypes stained with anti-PAR antibodies and DAPI. Right: PAR staining. Scale bar 20 μm. Analysis was performed in biological duplicates. b Western blot with fractionated extracts shows higher PARG-1CD::GFP and PARG-1CD abundance compared to controls. Anti-histone H3 was used as loading control for chromatin-bound fraction. Western blot (bottom) confirmed accumulation of PAR in both parg-1(cd) and parg-1(gk120) null mutants. Analysis was performed in biological duplicates. c Non-deconvolved images of late pachytene nuclei showing SYP-1 and elevated levels of PARG-1CD::GFP versus PARG-1::GFP with delayed redistribution in late pachytene nuclei. Scale bar 10 μm. Analysis was performed in biological duplicates. d The X-chromosome undergoes desynapsis in parg-1(cd); him-5 double mutants. Arrows indicate the unsynapsed X chromosome in the indicated genotypes before irradiation. Scale bar 10 μm. Analysis was performed in biological duplicates. e Exposure to IR fully rescues X-chromosome desynapsis in parg-1(cd); him-5 double mutants. Scale bar 10 μm. Analysis was performed in biological duplicates. f Quantification of synapsis by SYP-1 and HTP-3 co-staining in the indicated genotypes before and after IR exposure. Error bars indicated S.E.M. Statistical significance was assessed by χ2 test (significance level for p < 0.05, ***p < 0.0001, ns = not significant). Number of nuclei analysed (0 Gy-10 Gy): WT (215–205), parg-1(cd) (137–146), him-5 (169–190), parg-1(cd); him-5 (165–212). g Blocking PARG-1 catalytic activity caused milder synergistic effects when combined with him-5 mutants in contrast to parg-1(gk120); him-5. Center of error bars indicates mean. Error bars indicate standard deviation and asterisks show statistical significance calculated by two-tailed nonparametric Mann–Whitney test. (***p < 0.0001). Number of animals scored: parg-1::GFP (5), parg-1(gk120) (5), parg-1(cd)::GFP (4), him-5 (5), parg-1::GFP; him-5 (5), parg-1(gk120); him-5 (9), parg-1(cd); him-5 (10), parg-1(cd)::GFP; him-5 (10). h DAPI-staining and quantification of DAPI-bodies (i) in diakinesis nuclei for the indicated genotypes. Center of error bars indicates mean. Error bars indicate standard deviation and asterisks show statistical significance calculated by two-tailed nonparametric Mann–Whitney test. (***p < 0.0001). Scale bar 5 μm. Number of nuclei analysed is reported in Supplementary Table 2.