Figure 2. Strategy to mine mechanisms of natural stress resilience for novel antidepressant drug discovery.
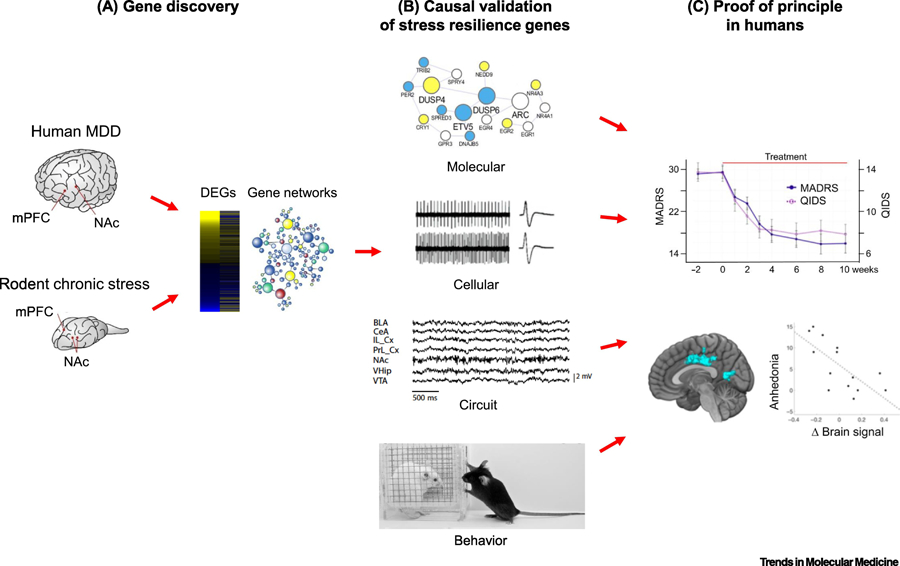
(A) Gene discovery involves using RNA-sequencing or other unbiased, genome-wide approaches to obtain an open-ended view of gene expression changes found in postmortem brains of humans with Major depressive disorder (MDD) or other stress-related disorders and in rodent chronic stress models. The latter include chronic social defeat stress, chronic variable stress, and paradigms of early life stress (e.g., maternal separation). Differentially expressed genes (DEGs) and key driver genes in gene networks are identified and correlated with resilience in rodents, and opposite effects in depressed humans. (B) A causal role of these key drivers in mediating resilience is then validated across molecular, cellular, circuit, and behavioral levels of analysis in rodent models. (C) Small molecules that affect a key driver can be studied in humans as a proof of principle, combining behavioral analyses with brain imaging measures. In the example shown, ezogabine decreases symptoms of depression in adult depressed humans based on the MADRS and QIDS behavioral rating scales: anhedonia measures within these rating scales correlate with a change in BOLD signal by functional MRI in the patients’s nucleus accumbens. (Data from [56]).
