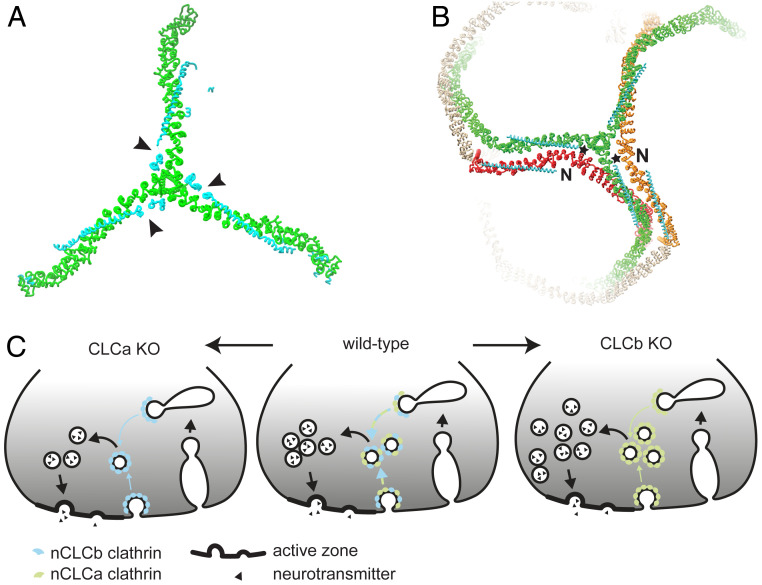
Fig. 6.
Model of how neuronal CLC diversity regulates SV recycling and lattice properties. (A) CLC (cyan) neuronal splice inserts (arrowheads) are located near the TxD of the bound CHC (green, left, PDB ID code 3LVH). (B) CLC (cyan) neuronal splice inserts (stars) are near the CHC knee of the neighboring triskelion (red or orange) within lattices (right, PDB ID code 3IYV). Interaction between a neuronal splicing insert and the knee of an adjacent triskelion and/or adjacent CLC N terminus (N) could promote conformational change in the CHC knee. As the formation of pentagons and hexagons requires different knee angles, this interaction could consequently influence lattice curvature (see Fig. 1A). (C) In WT mice (Center), a mix of nCLCa and nCLCb clathrin creates the appropriate biophysical properties to mediate SV generation from endosomal compartments (and possibly the plasma membrane, blue-green arrows). Loss of nCLCa (Left) creates nCLCb clathrin lattices that are defective in efficient SV regeneration (blue arrows), resulting in decimated SV pools. Loss of nCLCb (Right) creates nCLCa clathrin lattices, which are also less efficient in SV regeneration than WT, but able to maintain an overall increased SV pool by excess budding to compensate for reduced efficiency in acute SV pool replenishment (green arrows).