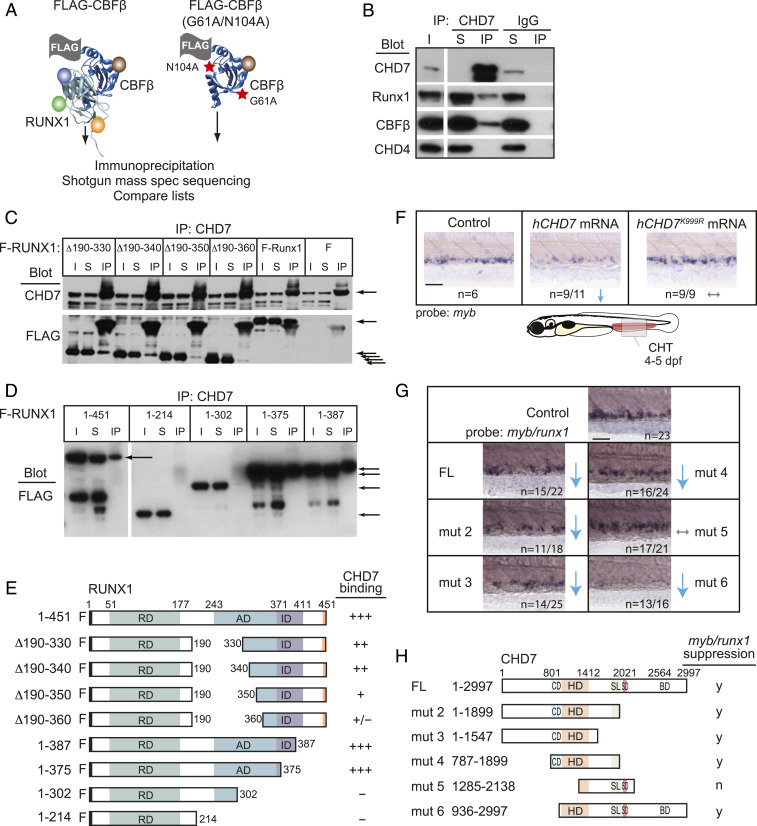
Fig. 4.
CHD7 interacts with Runx1 and restrains RUNX1 activity. (A) Scheme for identifying RUNX1-CBFβ–interacting proteins in a murine T-ALL cell line. FLAG-tagged CBFβ containing two amino acid substitutions (red stars) that decrease RUNX1 binding was used as a negative control. (B) CHD7 coimmunoprecipitates RUNX1-CBFβ but not CHD4 in murine T-ALL cells. I, input; S, depleted supernatant following immunoprecipitation; IP, immunoprecipitate. (C) Deletions impinging on the RUNX1 activation domain decrease the interaction between RUNX1 and CHD7. CHD7 was immunoprecipitated, and Western blots were probed with antibodies to CHD7 or FLAG. F-RUNX1, FLAG-RUNX1; ∆ refers to deleted amino acids illustrated schematically in panel E; F, vector expressing FLAG alone. Arrows indicate CHD7 (Top) or full-length and internally deleted RUNX1 proteins (Bottom). (D) C-terminal RUNX1 deletions. (E) Summary of RUNX1 mapping experiments. RD, DNA- and CBFβ-binding Runt domain; AD, transactivation domain; ID, inhibitory domain. (F) Expression of hCHD7 but not the catalytically dead mutant hCHD7K999R in zebrafish embryos reduces myb expression in the CHT by whole-mount in situ hybridization. Representative embryos are shown. Blue arrows indicate a decrease. Gray arrows indicate no change. (Scale bars, 50 μm.) Replicates: 2. (G) Mutation mapping of the hCHD7 domains shows that the ATPase/helicase domain is required to suppress myb and runx1 expression in the CHT. Same symbols as in F. (H) Summary of hCHD7 mapping experiments. FL, full length. CD, chromodomain. HD, ATPase/helicase domain. SL/SD/BD, SLIDE/SANT/BRK domains. y, yes; n, no. Quantification of results from F and G are in SI Appendix, Fig. S2B.