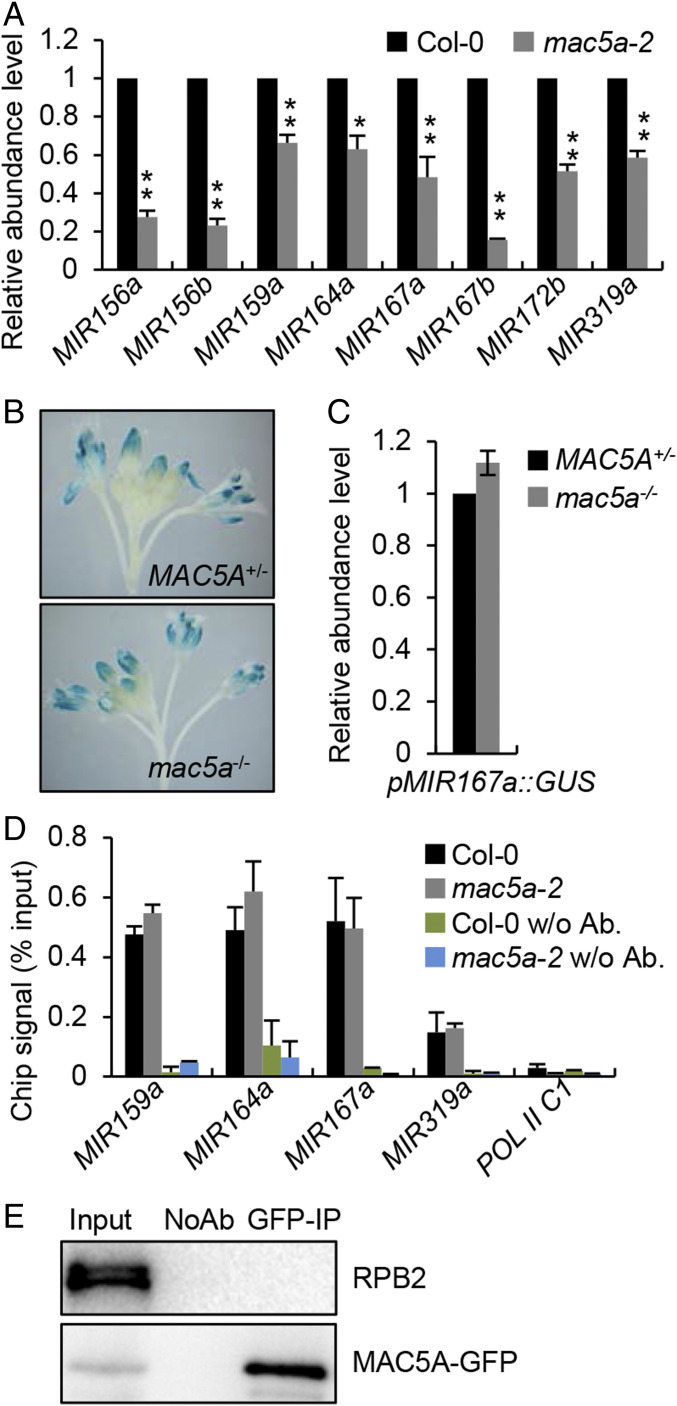
Fig. 2.
mac5a reduces pri-miRNA accumulation without affecting transcription. (A) qRT-PCR analysis of pri-miRNA levels in Col-0 and mac5a-2. pri-miRNA levels were normalized to those of UBQ5 and the values in Col-0 were set as 1. Error bars indicate SD of three replicates (*P < 0.05 and **P < 0.01 by Student’s t test compared with Col-0 values). (B) Histochemical staining of GUS in inflorescence of wild-type and mac5a-2 harboring pMIR167a::GUS transgene. Fifteen plants for each genotype were analyzed and a picture is shown. (C) Detection of relative levels of GUS transcript in the indicated plants (the values in Col-0 were set as 1). Error bar indicates SD of three replicates. (D) Pol II occupancy at MIR promoter in the indicated plants detected by ChIP followed by qPCR. The intergenic region between At2g17470 and At2G17460 (POL C1) was used as a negative control. (E) Co-IP between RPB2 and MAC5A. Protein extracts from inflorescences of gMAC5A-GFP transgenic plants were performed with anti-GFP antibodies. The proteins were detected by anti-GFP or anti-RRB2 antibodies. Input indicates total proteins before immunoprecipitation; NoAb indicates immunoprecipitation using agarose beads without antibodies.