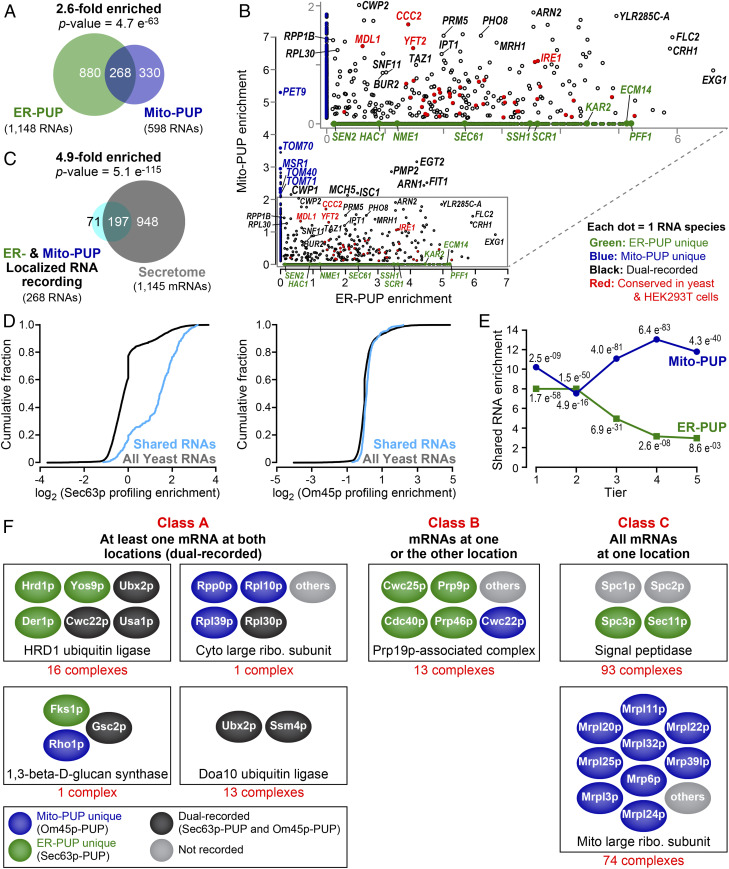
Fig. 4.
Dual recording at both ER and mitochondria. (A) mRNAs recorded by ER-PUP and Mito-PUP. (B) Comparison of RNAs uniquely enriched by the ER- (green dots) or Mito-PUP (blue dots), and those that were common to both (black, and red). Larger circles represent mRNAs that encode proteins typical of each organelle, while the red denote mRNAs that are detected at the ER (ER-PUP unique) and mitochondria (Mito-PUP unique) by independent, APEX-seq studies in HEK293T cells (24). (C) Dual-recorded RNAs vs. secretome mRNAs. (D) Cumulative fraction of common RNAs that have ER-profiling (20) (Left) and mitochondrial profiling (21) (Right) enrichment. (E) Distribution of common RNAs among ER- (green line) and Mito-PUP (blue line) rank. (F) mRNAs that encode subunits of multiprotein complexes exhibit three localization patterns. Representative complexes depicted for each class; physical interactions between proteins not implied. For a complete list, see Dataset S24.