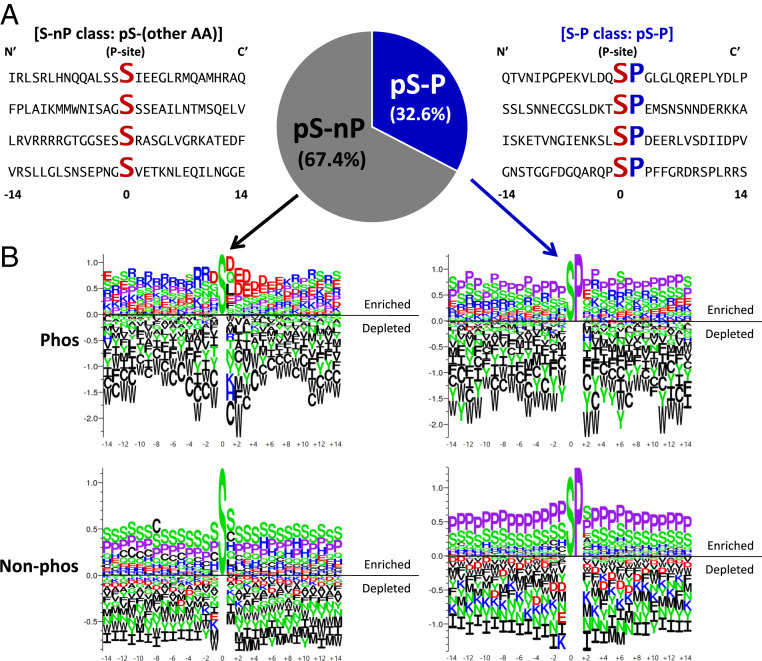
Fig. 3.
Proline residue at the +1 site (+1 Pro) of Ser phosphorylation sites defines a subclass of site (S-P) dependent on horizontal information. (A) Example 29-mer sequence neighborhoods centered on the phosphorylated Ser residue. Conserved Ser (S) and +1 Pro residues (P) are enlarged and bold. Frequencies of +1 Pro phosphorylation sites (S/T-P) make up one-third of all known human phosphorylated Ser. (B) Amino acid frequencies around S-P and S-nP demonstrate that S-P sites have little distinguishing sequence features as compared to nonphosphorylated sites with S-P dipeptide. (Top) Logos show enrichment/depletion patterns of amino acids around phosphorylated Ser sites. (Bottom) Logos show patterns around nonphosphorylated Ser sites. (Left) Logos show patterns where the Ser is immediately followed by amino acids other than Pro. (Right) Logos show patterns where the Ser is immediately followed by Pro (i.e., +1 Pro). Vertical scale indicates information content in bits. In all panels, aliphatic/nonpolar residues are colored black, prolines are lavender, polar residues are green, negatively charged side chains are red, and positively charged side chains are blue.