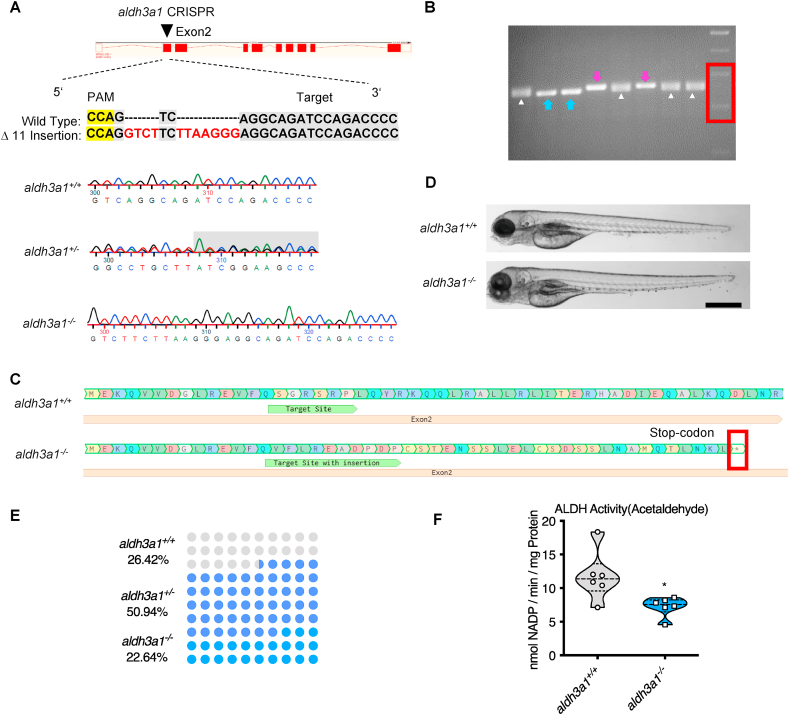
Fig. 2.
Generation of Aldh3a1 knockout zebrafish by using CRISPR-Cas9 technology.
(A). Aldh3a1-CRISPR-target site was designed in exon2 of aldh3a1 and CRISPR/Cas9-induced insertions of 11 nucleotides was selected for further investigation. Genotype was analysed and chromatogram showed aldh3a1 wild type, heterozygous, and homozygous sequencing results. (B). Genotyping-PCR gel showed the distinguishable bands of aldh3a1 wild type, homozygous and heterozygous zebrafish mutant line. The blue arrows, purple arrows and white deltas indicate the genotyping-PCR products from aldh3a1 wild type, homozygous and heterozygous zebrafish, respectively. The red frame indicates the marker, lower band is 200 bp and upper band is 300 bp. PCR product size: wildtype, 255 bp; homozygous, 266 bp. (C). Amino acid sequence showed a stop-codon in exon2 of aldh3a1−/−. (D). Microscopic images showed normal gross morphology of aldh3a1−/− larvae in comparison with aldh3a1+/+ larvae at 96 hpf. Black scale bar: 500 μm. (E). Adult fish number among different genotypes was in line with the Mendelian Inheritance in the first generation of F2: aldh3a1+/+ = 14, aldh3a1+/− = 27 and aldh3a1−/− = 12. (F). Aldh3a1−/− zebrafish showed decreased enzyme activity (acetaldehyde as substrate) measured by spectrophotometric analysis in zebrafish lysates at 96 hpf; n = 6 clutches with 46–50 larvae. For statistical analysis Student's t-test was applied, *p < 0.05. Bp, base pair. PAM, protospacer-adjacent motif. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)