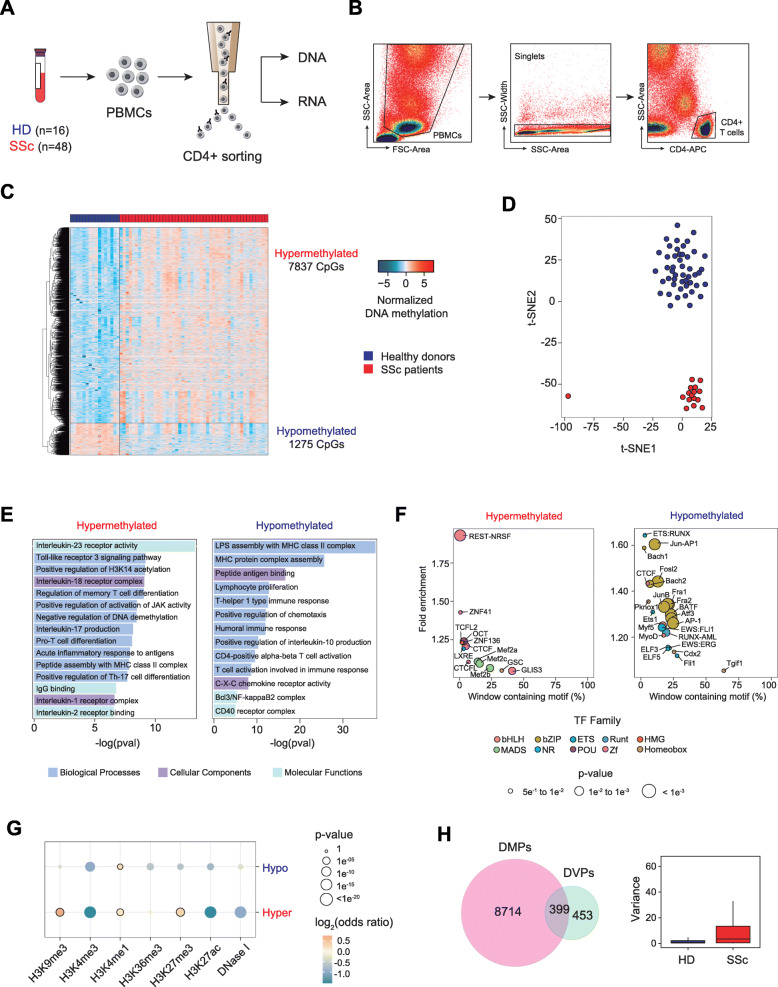
Fig. 1.
Aberrant DNA methylation in inflammatory loci of SSc CD4+ T cells. a Scheme depicting workflow including PBMC isolation, CD4+ T cell positive sorting, and DNA and RNA extraction. b Gating strategy to eliminate cell debris, doublets, and the isolation of CD4-APC+ T lymphocytes by side scatter. c Heatmap of differential methylated CpGs (DMPs), in which 7837 CpGs were hypermethylated (log2FC > 0, FDR < 0.05) and 1275 CpGs were hypomethylated (log2FC < 0, FDR < 0.05) in SSc patients (n = 48) compared to healthy control (n = 16). d t-SNE clustering of SSc and HD samples based on all identified DMPs. e Gene ontology of hyper- and hypomethylated DMPs, analyzed utilizing the GREAT online tool (http://great.stanford.edu/), in which CpGs annotated in the EPIC array were used as background. f HOMER motif enrichment of hyper- and hypomethylated CpGs, utilizing CpGs annotated in the EPIC array as background. A window of ± 250 bp centering around each DMP was applied. g Enrichment of DNase I hypersensitivity, H3K9me3, H3K4me3, H3K4me1, H3K36me3, H3K27me3, and H3K27ac ChIP-seq data, obtained from the BLUEPRINT portal, in hyper- and hypomethylated DMPs. Highlighted circles represent statistically significant comparisons (p value < 0.01 and odds ratio > 1) compared to background. h Overlap between DMPs and differentially variable CpGs (DVPs), identified utilizing the iEVORA algorithm (left), and graphical representation of variance of identified DVPs in HD and SSc