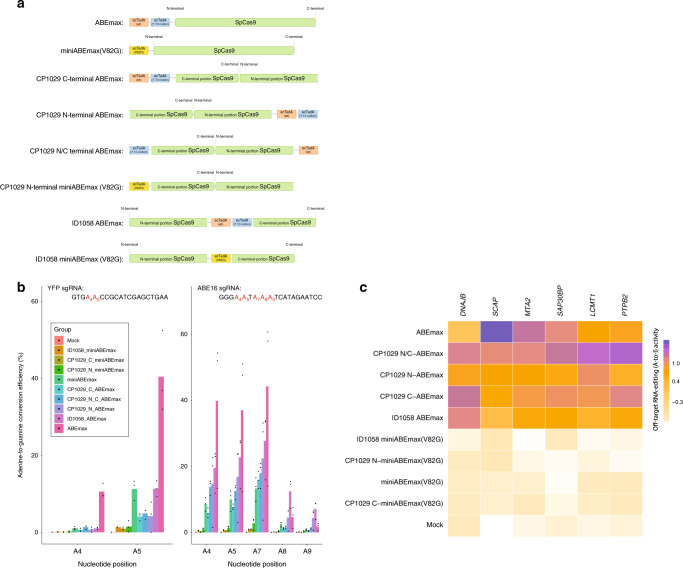
Fig. 2. Activity profile of nSpCas9-adenine base editor (ABE) permutations.
a The wild-type TadA and the evolved TadA monomer for ABEmax are light blue and orange stadiums, respectively, while the miniABEmaxV82G is depicted as a yellow stadium. b On-target activity profile of different ABE permutants tested. nSpCas9-variants of ABE permutants were challenged against the YFP and ABE16 loci. Data are pooled from three independent technical replicates, showing the mean (colored bar) activity at several adenine positions measured for adenine-to-guanine editing. c Heatmap showing the off-target profile of different permutations of nSpCas9 ABEs. Off-target events were considered at the position of the most heavily deaminated adenine in the promiscuous DNAJB, SCAP, MTA2, SAP30BP, LCMT1, and PTPB2 transcripts (three independent technical replicates of three biological replicates targeting YFP, ABE16, and non-targeting were included). Adenosine (A) to inosine (I) editing efficiencies are shown as the mean value scaled from the lowest to highest value across all samples for all RNA off-target sites. Source data are available in the Source data file.