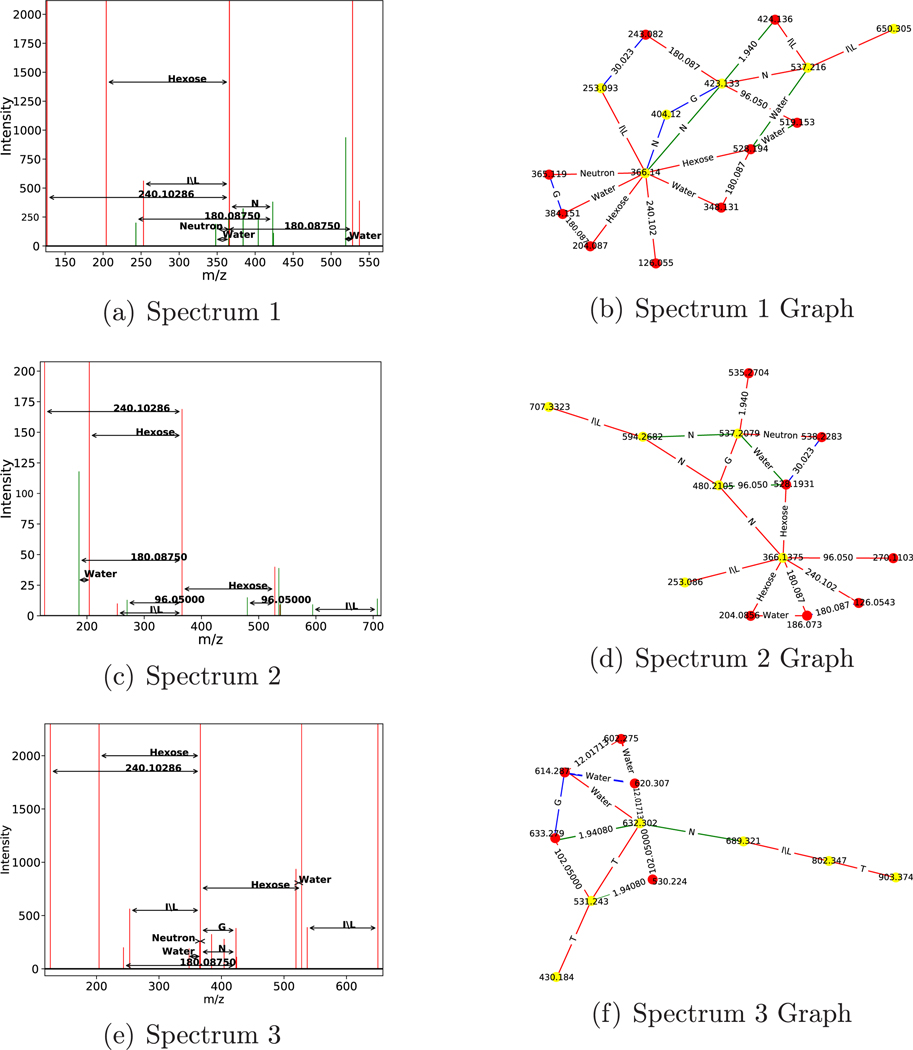
Figure 6.
Subgraphs with an amino acid chain matching glycosylation sites. Three spectra (a,c,e) and their corresponding de novo graphs (b,d,f) found using the combinatorial approach. The top two spectra contain the amino acid chain LNGNL, and the bottom contains the amino acid chain TLNTT. Graphs use red edges to mark charge z = 1, green edges to mark z = 2, and blue edges to mark z = 3. The nodes colored yellow represent nodes touched by the amino acid chain. Panels a and b came from spectrum titled “Locus:1.1.1.8405.3”. Panels c and d came from spectrum titled “Locus:1.1.1.8036.2”. Panels e and f came from “Locus:1.1.1.2523.2”.