ABSTRACT
Mammalian circadian rhythms drive ∼24 h periodicity in a wide range of cellular processes, temporally coordinating physiology and behaviour within an organism, and synchronising this with the external day–night cycle. The canonical model for this timekeeping consists of a delayed negative-feedback loop, containing transcriptional activator complex CLOCK–BMAL1 (BMAL1 is also known as ARNTL) and repressors period 1, 2 and 3 (PER1, PER2 and PER3) and cryptochrome 1 and 2 (CRY1 and CRY2), along with a number of accessory factors. Although the broad strokes of this system are defined, the exact molecular mechanisms by which these proteins generate a self-sustained rhythm with such periodicity and fidelity remains a topic of much research. Recent studies have identified prominent roles for a number of crucial post-transcriptional, translational and, particularly, post-translational events within the mammalian circadian oscillator, providing an increasingly complex understanding of the activities and interactions of the core clock proteins. In this Review, we highlight such contemporary work on non-transcriptional events and set it within our current understanding of cellular circadian timekeeping.
KEY WORDS: Circadian rhythm, Post-transcriptional modification, Post-translational modification, Translational regulation, Cellular timekeeping
Summary: In this Review, we highlight recent work identifying non-transcriptional mechanisms crucial to regulating the core mammalian clock proteins and their role in circadian rhythmicity.
Introduction
We live in a 24-h world, where the rotation of the Earth provides a continuous environmental rhythm – the day–night cycle. Consequentially, animals, plants, fungi and some bacteria (Dunlap, 1999; Roenneberg and Merrow, 2005) have evolved to exhibit circadian rhythms in a wide range of biological processes, from sleep–wake cycles to wound healing (Hoyle et al., 2017). These rhythms exhibit an ∼24-h period, meaning that the time taken for each cycle is around a day. They are self-perpetuating, persisting without external timing information, but are able to shift their phase of oscillation to synchronise with specific external cues, such as the light–dark cycle, feeding and temperature. Furthermore, they are temperature compensated, retaining their 24-h periodicity across the range of biologically relevant temperatures (Pittendrigh, 1960). Circadian rhythmicity is thought to provide a significant fitness advantage (DeCoursey et al., 2000; Woelfle et al., 2004), as it allows organisms to predict the timing of regular external events, modifying their physiology and behaviour in anticipation of environmental change, rather than in response to it. Furthermore, circadian disruption, as occurs during shift work, where people are active when their physiology would favour sleep, is associated with adverse health outcomes, including increased rates of type 2 diabetes, some forms of cancer, and compromised immune function (Cuesta et al., 2016; Salgado-Delgado et al., 2013; Scheer et al., 2009). Conversely, disrupted circadian behaviour and physiology is an early symptom of some neurological disorders, including Parkinson's and Alzheimer's disease (Li et al., 2017; Saeed and Abbott, 2017). Thus, maintenance of circadian rhythmicity is closely associated with long-term human health (Reddy and O'Neill, 2010).
Mammalian circadian rhythms have a cell-autonomous basis (Balsalobre et al., 2000; Welsh et al., 2005; Yoo et al., 2004), thought to be driven by a transcriptional–translational feedback loop (TTFL). At its heart, this contains transcriptional activators circadian locomotor output cycles protein kaput (CLOCK) and brain and muscle ARNT-like 1 (BMAL1; also known as ARNTL), which together form an active complex, and repressors period (PER; of which there are three isoforms, PER1, PER2 and PER3) and cryptochrome (CRY; of which there are two isoforms, CRY1 and CRY2; see Box 1), collectively referred to here as ‘core clock proteins’. These are further regulated by an accessory loop containing the nuclear receptors Rev-Erbα and Rev-Erbβ (also known as NR1D1 and NR1D2, respectively) and retinoic acid receptor-related orphan receptors (RORs, consisting of RORα, -β and -γ; also known as RORA–RORC), which drive rhythmic expression of BMAL1. This accessory loop, unlike the core clock proteins, is not essential for maintenance of fully-fledged circadian rhythmicity, but does play a role in regulating rhythmic expression of downstream targets or ‘clock-controlled genes’, which have circadian changes in expression, but do not regulate circadian timekeeping themselves (Ikeda et al., 2019; Liu et al., 2008).
Box 1. An overview of the core mammalian molecular circadian oscillator.
The canonical model of cellular circadian timekeeping centres around a negative transcriptional-translational feedback loop (TTFL) (Takahashi et al., 2017). Here, the basic helix-loop-helix (bHLH) PER-ARNT-SIM (PAS) proteins CLOCK and BMAL1 heterodimerise and bind to E-boxes (a DNA response element) to drive the transcription of a large number of genes, including the period (PER) proteins PER1, PER2 and PER3, and the cryptochrome (CRY) proteins CRY1 and CRY2, which are transcriptional repressors of CLOCK-BMAL1. Following translation, both repressors form a complex, together with CK1, and translocate to the nucleus. Here they repress the activity of CLOCK–BMAL1 at E-box-regulated genes through the formation of a macromolecular assembly, containing CLOCK, BMAL1, PER, CRY and CK1 (Aryal et al., 2017) (see figure). Nascent PER and CRY expression subsequently drops, and existing PER and CRY proteins are targeted for ubiquitylation and subsequent degradation by E3 ligase complexes. Both PER and CRY have relatively short half-lives; thus, when their transcription is inhibited, their relative abundance drops quickly. This reduces the abundance of the repressive complex, and so relieves repression of CLOCK–BMAL1 activity. CLOCK–BMAL1 is therefore able to resume its transcriptional activity at E-boxes, thus restarting the loop. This entire oscillation takes ∼24 h (Takahashi, 2017).
CLOCK–BMAL1 also drives expression of the nuclear receptors Rev-Erbα and Rev-Erbβ, while BMAL1 expression is driven by the binding of RORα and RORβ to ROR-response elements in the BMAL1 promoter (see figure). Rev-Erb proteins inhibit the activity of RORs on BMAL1 expression, and also inhibit transcription of Clock (Crumbley and Burris, 2011). In turn, expression of RORα and RORβ is driven by D site-binding protein (DBP) and Nfil3 binding to D-boxes in the ROR promoter region. Together, this forms a secondary transcriptional-translational feedback loop (Takahashi, 2017). Although this is important for driving the circadian expression of a number of downstream targets, this accessory loop is not thought to be as crucial for maintaining cellular timekeeping as the core loop containing CLOCK, BMAL1, PER and CRY (Ikeda et al., 2019; Liu et al., 2008).
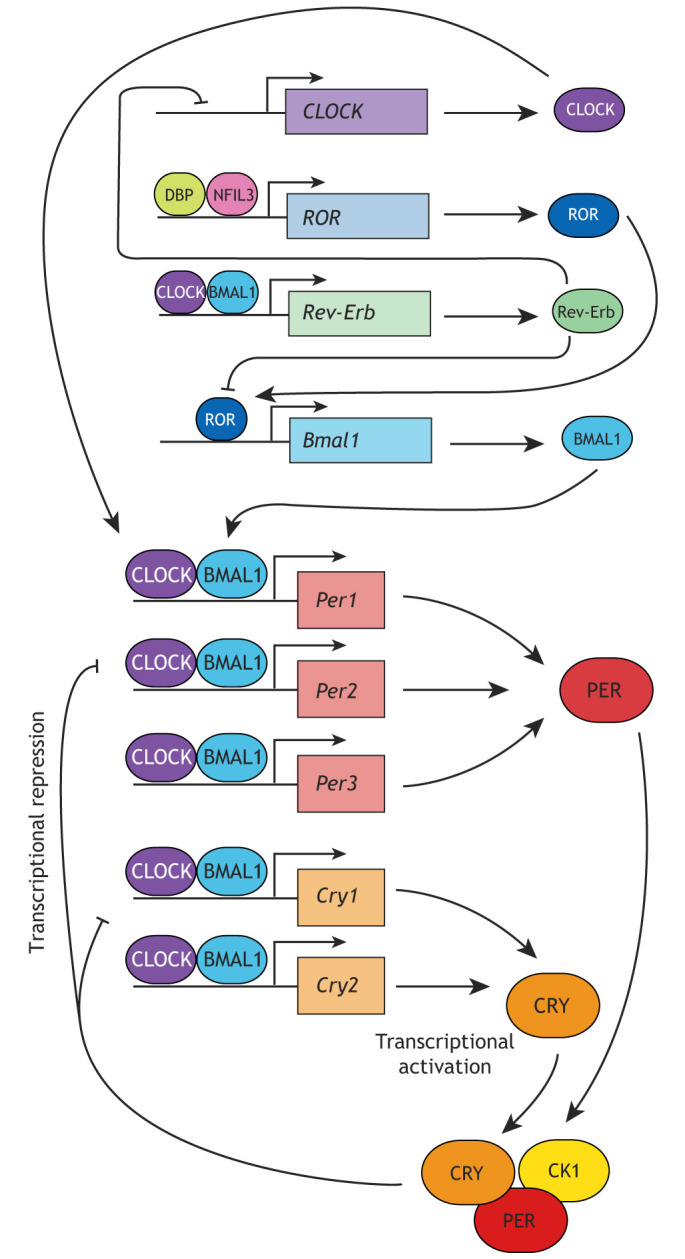
In the past 20 years, much attention has been paid to the transcriptional function of the core clock proteins and their targets. Such work has recently been excellently and extensively reviewed (Takahashi, 2017), and so will not be further discussed here, but a brief introduction to this core TTFL can be found in Box 1.
Increasingly, however, it is apparent that expression and activity of clock proteins is also regulated by non-transcriptional processes, which are crucial for maintaining circadian timekeeping over and above the transcriptional framework. Here, we highlight recent work concerning these non-transcriptional mechanisms, and discuss how such findings extend our existing understanding of the molecular mechanisms of mammalian timekeeping.
Post-transcriptional regulation of ‘clock gene’ transcripts
Following transcription, mRNA can be extensively regulated and modified before being translated (Fabian et al., 2010; García-Mauriño et al., 2017; Leppek et al., 2018; Zhao et al., 2017), and circadian transcripts are no exception (Fig. 1). Around 40% of the transcriptome oscillates in at least one tissue (Anafi et al., 2017; Zhang et al., 2014), However, there is discrepancy between those transcripts with oscillatory abundance, and those with an oscillation in nascent transcription, with only ∼25% of rhythmically abundant RNAs being rhythmically transcribed, thus indicating significant roles for rhythmic regulation and degradation of transcripts (Koike et al., 2012; Lück et al., 2014; Menet et al., 2012). That said, the so-called ‘core clock’ genes are generally rhythmically transcribed, with ribosome occupancy of those transcripts in the same phase as their transcription (Janich et al., 2015; Koike et al., 2012; Rey et al., 2011), indicating that rhythmic transcription does regulate rhythmic expression of these proteins.
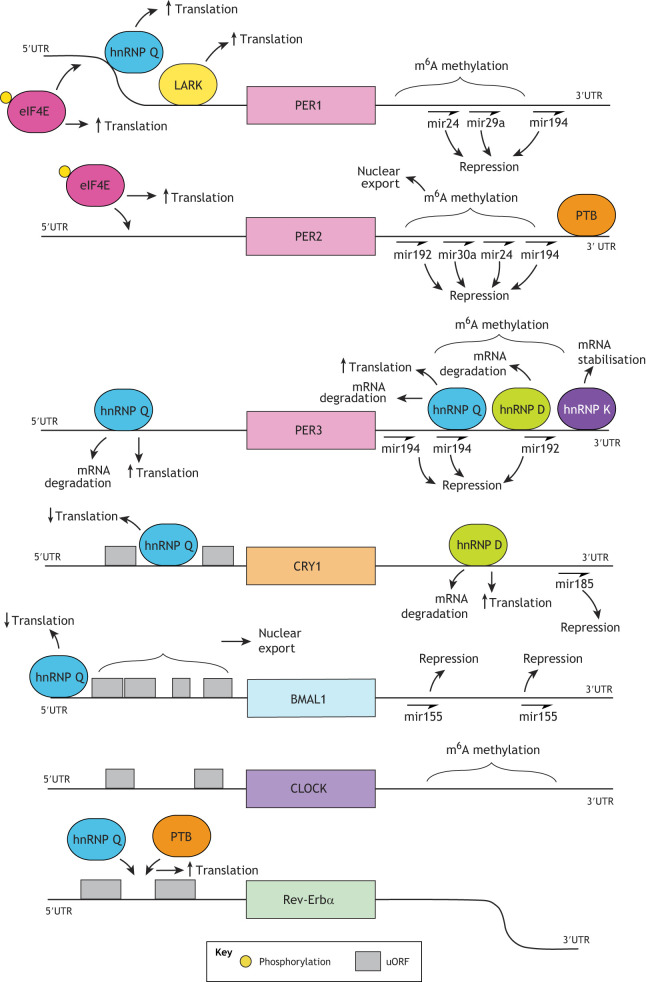
Fig. 1.
Schematic overview of factors regulating mRNA levels of core clock proteins. Myriad factors influence the stability of mRNA coding for core clock proteins. Coding sequences are represented as coloured blocks, flanked by 5′ and 3′ UTRs. Large coloured lozenge shapes represent proteins and half-arrows denote miRNA binding. Readers should be aware that the exact binding sites of some proteins are not currently strictly defined, and so their order relative to each other is not definitive.
There is, however, clear evidence for post-transcriptional regulation in determination of circadian periodicity. One study identified selective N6 methylation of adenosine (m6A) in Per1, Per2 and Per3, Clock, Bmal1 and Rev-Erbα transcripts (Fig. 1), and showed that m6A methylation of Per2 and Bmal1 (and likely other transcripts) aids their nuclear export (Fustin et al., 2013). Inhibition of methylation lengthened circadian period, whereas overexpression of the Mettl3 methylating enzyme shortened the period (Fustin et al., 2013). This group also showed that 3′ untranslated region (UTR) m6A methylation negatively regulates the expression of casein kinase 1 (CK1)δ splice variants CK1δ1 and CK1δ2, with removal of this methylation site simultaneously increasing Csnk1d1 (encoding CK1δ1) mRNA levels and increasing translation of both variants, as well as lengthening circadian period in mice (Fustin et al., 2018). The same team suggest a role for m7G-cap methylation in nuclear export of core clock transcripts, but whether this is a ubiquitous process, or has some sequence specificity, remains unclear (Fustin et al., 2013). Both Clock and Rev-Erbα mRNA also undergo alternative splicing, with splice variants showing a different phase of expression relative to the main transcript (McGlincy et al., 2012). However, the function(s) of these variants is not currently well understood.
The 3′UTR plays a particularly important role in regulating the stability and degradation of core clock transcripts (Fig. 1). Per2 transcripts lacking their native 3′UTR have a longer half-life than those containing it (Woo et al., 2009), and the Per3 3′UTR also substantially promotes mRNA degradation (Kwak et al., 2006). Similarly, replacing the Per2 3′UTR with the SV40 late poly(A) signal in mice produced a longer free-running period and concomitantly increased cellular period and amplitude in various tissues, suggesting that endogenous 3′UTR elements promote Per2 mRNA degradation (Yoo et al., 2017). This has been partly attributed to polypyrimidine tract-binding protein (PTB, herein referring to PTBP1), which binds to the Per2 3′UTR to promote Per2 mRNA degradation (Woo et al., 2009). Furthermore, expression of PTB oscillates in antiphase with Per2 mRNA, with PTB levels increasing as Per2 mRNA levels decrease (Woo et al., 2009). Thus, Per2 mRNA degradation via PTB is increased at the end of the repressive phase (see Box 1). PTB-mediated destabilisation of Per2 mRNA during this phase further reduces the potential for production of nascent PER2 protein, aiding the end of transcriptional repression by PER2.
Cry1 mRNA is also regulated by its 3′UTR, with the presence of this UTR associated with reduced mRNA levels (Lee et al., 2014). This is partially attributed to heterogeneous nuclear ribonucleoprotein D/AU-rich element (ARE) RNA-binding protein 1 (hnRNP D, also known as AUF1), which binds to an ARE within the Cry1 3′UTR, promoting Cry1 mRNA degradation (Lee et al., 2014; Woo et al., 2010). As with PTB, cytoplasmic expression of hnRNP D oscillates in the opposite phase to Cry1 mRNA. Per3 mRNA stability is also reduced by binding of either hnRNP D to its 3′UTR or hnRNP Q to both its 5′ and 3′UTRs, while binding of hnRNP K to the Per3 3′UTR has a stabilising effect (Kim et al., 2011, 2015) (Fig. 1).
The 3′UTR is also a significant site of regulation by micro (mi)RNAs. It was observed over a decade ago that the miR192/4 cluster regulated all three Per transcripts at their 3′UTR (Nagel et al., 2009). The Lee laboratory identified three further miRNAs (miR-24, miR29a and miR30a) that target the Per1 and Per2 3′UTR (Chen et al., 2013). Knockdown of these miRNAs, or overexpression of their 3′UTR targets, led to an ∼90 min period shortening (Chen et al., 2013), with miR-24 alone posited to be sufficient to destabilise Per2 mRNA (Yoo et al., 2017). Selective downregulation of these Per-targeting miRNAs is important in synchronisation of cellular circadian rhythms to feeding (Crosby et al., 2019).
miR-155 regulates the Bmal1 3′UTR within the innate immune system; an immune challenge using lipopolysaccharide, a pro-inflammatory cell-wall component of gram-negative bacteria, leads to increased miR-155 and a subsequent decrease in both Bmal1 mRNA and protein levels (Curtis et al., 2015). Meanwhile, miR-185 represses Cry1 translation through interaction with its 3′UTR, with cytoplasmic expression of this miRNA in antiphase with the abundance of CRY1 protein (Lee et al., 2013).
Translational regulation of ‘clock protein’ production
Lack of circadian oscillation in the abundance or modification of a transcript does not necessarily imply a lack of oscillation in the abundance of the related protein. Work on circadian changes in the proteome suggests that although ∼20% of soluble proteins in mouse liver oscillate, only around half of these have a corresponding oscillatory transcript (Mauvoisin et al., 2014; Reddy et al., 2006; Robles et al., 2014).
The explanation for this may lie in translational control. Previous work has reported circadian variation in translational efficiency, with a greater proportion of ribosomes in large polysomes, and greater cellular protein content, during the active phase compared to the resting phase (Jouffe et al., 2013; Sinturel et al., 2017). This could lead to a model where a subset of non-rhythmic transcripts exhibit rhythmic protein expression as a result of circadian changes in ribosome availability and activity. The specificity of such rhythmic translational activity remains to be explored; transcripts containing either a 5′-terminal oligo pyrimidine tract (5′TOP) or translation initiator or short 5′UTR (TISU) motif are enriched in the group of transcripts that show rhythmic changes in translation without rhythmic transcription (Atger et al., 2015; Jouffe et al., 2013). However, there are constantly expressed transcripts with rhythmic translation that do not contain these motifs (Atger et al., 2015; Jang et al., 2015), making this an exciting area for future work.
To date, the contribution of circadian variation in translational efficiency to the expression of the core clock proteins is unclear; they do not contain identified TOP or TISU sequences. There is, however, compelling evidence supporting differential translational regulation between the core clock proteins, resulting in non-circadian differences in translational efficiency. For instance, ribosome occupancy has been shown to vary up to 6-fold between core clock transcripts (Janich et al., 2015) such that, although overall mRNA levels vary between clock gene transcripts, total nascent translation of each protein is comparable to that of the other core clock proteins. Variable ribosome occupancy is driven in part by the presence of upstream open reading frames (uORFs) in the 5′UTR; Bmal1, Clock, Cry1 and Rev-Erbα and Rev-Erbβ all have at least one uORF, with ribosome occupancy at these uORFs capable of influencing translational efficiency of the main ORF (Jang et al., 2015; Janich et al., 2015) (Fig. 1). For example, uORF deletion in Rev-Erbα did not alter the period, but did produce increased mRNA stability and increased translation of the main ORF, thus increasing the amplitude of Rev-Erbα expression (Janich et al., 2015). Furthermore, decreasing reinitiation of translation between the uORF and the main ORF through knockdown of density regulated protein (DENR), which acts to selectively promote reinitation of translation at the main ORF after translational termination at an uORF (Schleich et al., 2014), resulted in period shortening of up to 90 min (Janich et al., 2015), hinting at further roles for uORFs in the maintenance of circadian period. Recent work suggests that Clock may be a particular target of DENR, as mutation of its two uORFs increased translational activity by more than 90% and abolished the DENR dependence of CLOCK translation (Castelo-Szekely et al., 2019). Intriguingly, in light of these uORFs, the authors propose an alternative AUG for Clock that is nine amino acids shorter than has been previously annotated, and demonstrate far higher translational activity from this alternative site than from the canonical AUG (Castelo-Szekely et al., 2019).
The 5′UTR of Per also regulates its expression, even in the absence of an uORF. Rhythmic phosphorylation of initiation factor eIF4E, a downstream effector of the MAPK pathway, selectively promotes PER1 and PER2 translation through an interaction with their 5′UTR at an unidentified motif (Cao et al., 2015). Tissues from knock-in mice with an eIF4E mutant that cannot be phosphorylated showed a selective reduction in PER1 and PER2 translation, with fewer ribosomes associated with these transcripts compared to what was observed in wild-type animals. In contrast, global translation, and translation of other clock proteins in the knock-in mouse tissues, remained comparable to that in wild type. Furthermore, knock-in mice showed a reduced capacity to shift their behavioural rhythms in response to lighting cues (Cao et al., 2015).
In addition to its previously discussed role in Cry1 mRNA degradation, hnRNP D upregulates CRY1 translation (Lee et al., 2014). Here, it is proposed, hnRNP D bound to the Cry1 3′UTR further interacts with both translation initiation factors and ribosomal proteins, particularly 40S ribosomal subunit components, promoting Cry1 translation (Lee et al., 2014). Coupled with increased Cry1 mRNA degradation, this dual activity of hnRNP D may result in more acute expression of CRY1. hnRNP Q is similarly suggested to influence mRNA stability and increase translation upon binding to both the 5′ and 3′UTRs of Per3 (Kim et al., 2011). By contrast, binding of hnRNP Q to the 5′UTR of Bmal1 and Cry1 reduces the amplitude of their translation (Jung et al., 2019; Lim et al., 2016). hnRNP Q also modulates expression of Per1 and Rev-Erbα, through association with an internal ribosome entry site (IRES) within their respective 5′UTRs. Here, hnRNP Q reduces the amplitude of PER1 expression, whereas knockdown of hnRNP Q, together with PTB knockdown, in NIH3T3 cells abolished rhythmic expression of Rev-Erbα without appreciably influencing its rhythmic RNA abundance (Lee et al., 2012a,b; Kim et al., 2010). Finally, LARK has been shown to promote cap-dependent translation of Per1, through interaction with its 3′UTR, with LARK overexpression modestly increasing cellular period (Kojima et al., 2007) (Fig. 1).
Another well-studied translational regulator, mammalian target of rapamycin (mTOR), plays a role in acute translation of CRY and PER proteins, but not CLOCK and BMAL1, following cues known to influence circadian timekeeping. These cues include feeding, where post-prandial insulin signalling and subsequent mTOR activation drives a selective increase in expression of the PER proteins (Crosby et al., 2019), and serum-shock, which drives an acute increase in CRY1 expression (Ramanathan et al., 2018), with both increases abolished in the presence of rapamycin. Furthermore, reducing mTOR activity, either pharmacologically or by siRNA knockdown, results in increased period in cells, whereas increased mTOR activity shortens period (Feeney et al., 2016; Ramanathan et al., 2018). Additionally, mice possessing a heterozygous mTOR deletion show a modest but significant increase in behavioural period (Ramanathan et al., 2018), and mice treated with rapamycin showed a reduced capacity to synchronise their behaviour with changes in light–dark schedule (Cao et al., 2010). It has been suggested that mTOR activity, which is magnesium sensitive, might be regulated in a circadian manner as a consequence of circadian changes in intracellular magnesium content, which is highest during the subjective day, when mTOR activity also peaks (Feeney et al., 2016). Here, the authors show that depletion of magnesium from the extracellular medium of both the marine green alga Ostreococcus tauri and a human-derived cell line results in period lengthening, and that simultaneous pharmacological inhibition of mTOR activity does not compound this effect, suggesting that the two operate through the same pathway (Feeney et al., 2016). Additionally, mTOR is thought to be critical for the translational regulation of TOP mRNAs, as discussed above, through an mTORC1-independent mechanism (Meyuhas and Kahan, 2015; Cao et al., 2010).
Post-translational modification is crucial in regulation of clock protein activity and function
Once translated, the core clock proteins are subject to an array of post-translational modifications (Figs 2 and 3). It is worth noting, however, that these proteins do not drive rhythmicity individually, but instead form a number of macromolecular complexes to carry out their function, such as the CLOCK–BMAL1 complex, which drives transcriptional activation, and the repressive complex, which brings CLOCK–BMAL1, CRY, PER and CK1 together in close proximity and inhibits transcription (Aryal et al., 2017). There is growing insight into exactly how core clock proteins interact with each other within these complexes. Although such work falls outside the scope of this Review, we direct interested readers to our recent review exploring this topic (Partch, 2020). Readers should be aware however, that many of the post-translational modifications described below only occur within, or are modulated by, the formation of specific complexes.
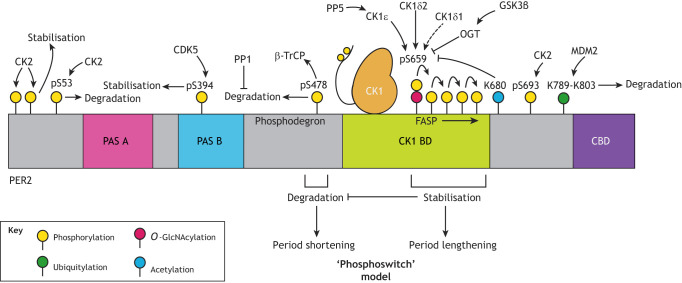
Fig. 2.
Schematic illustration of post-translational regulation of PER2. PER2 abundance and stability is a crucial determinant of circadian period and phase, with PER stabilisation associated with increased cellular period, whereas destabilisation of PER2 shortens the period. Phosphorylation of PER2 by CK1 is integral to regulation of PER2 stability. CK1δ and CK1ε bind to PER2 at the CK1-binding domain (CK1 BD) and can either stabilise or promote degradation of PER2, depending on the exact modification site, with phosphorylation at S478 in the phosphodegron promoting recruitment of the E3 ubiquitin ligase β-TrCP and thus degradation, whereas phosphorylation at the FASP region delays degradation. This interplay constitutes the ‘phosphoswitch’ model, which proposes that the balance of phosphorylation between these stabilising and degrading regions determines overall PER2 half-life. Dephosphorylation of PER2 by protein phosphatase 1 (PP1) reduces its overall degradation. In addition, CK2-mediated phosphorylation at the extreme N-terminus is proposed to stabilise the protein, whereas CK2 phosphorylation at S53 is thought to promote degradation, although more recent work suggests that the most significant site of CK2 phosphorylation on PER2 is S693. O-GlcNAcylation by O-GlcNAc transferase (OGT) at the priming serine (S659 in mice/S662 in humans) of the FASP site competitively inhibits PER2 phosphorylation by CK1, with GSK3β being a positive regulator of OGT activity. Acetylation at K680 also inhibits phosphorylation at the priming serine residue. Furthermore, ubiquitylation by MDM2 at sites downstream of FASP promotes PER2 degradation in a phosphorylation-independent manner, whereas phosphorylation at S394 within the PAS-B domain by CDK5 promotes stability. Phosphorylation and dephosphorylation (e.g. by PP5) of the C-terminal tail of CK1 can also influence its activity on its PER2 substrate. Further modifications that are discussed in the text but which currently lack a defined site of action or function, including direct phosphorylation of PER2 by GSK3β, decacetylation by SIRT1 and some O-GlcNAcylated serine residues, are not shown.
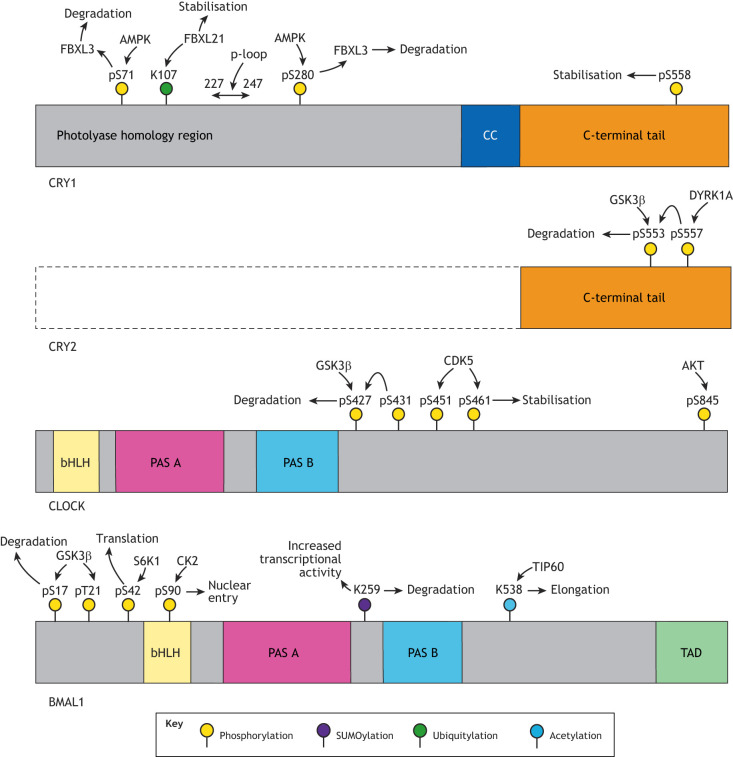
Fig. 3.
Schematic illustration overview of post-translational modifications on other core clock proteins. Although post-translational modifications of PER2 are the most studied, other core clock proteins are also controlled by significant post-translational modification. Only the C-terminal tail of CRY2 is shown, but the high similarity between the CRY1 and CRY2 photolyase-homology region (PHR) likely means that post-translational events applicable to CRY1 also occur on CRY2. Owing to their large number, only selected CRY1 phosphosites are shown.
A central role for CK1
Although the circadian field has historically focused on transcriptional mechanisms, there has long been appreciation of the crucial role played by some post-translational modifications. This is in no small part due to the fact that the first mammalian circadian mutant identified was in CK1, alterations in whose activity were found to result in an ∼20 h behavioural period (Lowrey et al., 2000; Ralph and Menaker, 1988). Despite this early indication, the precise mechanism through which CK1 regulates circadian period remains a fertile area for research.
CK1 has two relevant isoforms in mammals, CK1δ and CK1ε, although recent work suggests a greater role for CK1δ in regulating its major circadian target, the PER proteins (Lee et al., 2001; Meng et al., 2010, 2008). The most current model considers two regulatory regions on PER2 (Zhou et al., 2015) (Fig. 2). The first region, referred to as the ‘FASP’ region, is named after the discovery of an S662G mutation that results in the hereditary familial advanced sleep phase (FASP) syndrome (Vanselow et al., 2006; Toh et al., 2001). Phosphorylation of five sites within the FASP region increases PER stability – a non-consensus site at S659 in mice (S662 in humans), followed by four serine residues that conform to the pSXXS motif favoured by CK1 (Shanware et al., 2011). All of these sites are phosphorylated by CK1δ and/or CK1ε, with slow phosphorylation of the first, non-consensus serine being required to prime sequential phosphorylation of the following sites (Narasimamurthy et al., 2018). The second major site in PER2 that is regulated by CK1 is a region just after its PAS-B domain at S478 in mice (S480 in humans), referred to as the ‘phosphodegron’. Phosphorylation here prompts recruitment of the E3-ubiquitin ligase β-TrCP and subsequent PER2 degradation (Eide et al., 2005; Ohsaki et al., 2008; Reischl et al., 2007; Zhou et al., 2015). Recent work demonstrates that FASP phosphorylation decreases CK1 activity on the phosphodegron, although the exact mechanism underlying this remains unclear (Philpott et al., 2020; Zhou et al., 2015). Additionally, the two splice variants of CK1δ mentioned above, CK1δ1 and CK1δ2, have differing substrate preferences within PER2, with CK1δ2 having greater activity at the priming serine residue, thereby preferentially stabilising PER2 and promoting period lengthening (Fustin et al., 2018) (Fig. 2). Finally, phosphorylation of the C-terminal ‘tail’ of CK1, both through autophosphorylation and by a number of other kinases, also influences its activity for its substrate, with inhibition of this phosphorylation decreasing PER2 stability (Dahlberg et al., 2009; Eng et al., 2017; Giamas et al., 2007; Um et al., 2007). Together, the relative phosphorylation of the FASP and phosphodegron regions by CK1δ and/or CK1ε is currently thought to determine PER2 stability through the so-called ‘phosphoswitch’ model (Philpott et al., 2020; Zhou et al., 2015) (Fig. 2). As PER protein abundance is a crucial determinant of circadian period and phase (Chen et al., 2009), such activity of CK1 is therefore a major regulator of circadian rhythmicity.
Elegant as the phosphoswitch model is, it is likely that additional insights into the regulation of PER stability will require this model to become more complex. In particular, it should be noted that the S478 phosphodegron is not present in PER1, which possesses an alternative N-terminal phosphodegron motif (residues 121–126 in humans and mice) that also promotes the recruitment of β-TrCP to PER1 in a CK1-dependent manner (Ohsaki et al., 2008; Shirogane et al., 2005). This N-terminal motif is conserved in PER2, but its role in regulating PER2 degradation in addition to the degron proposed in the phosphoswitch model is unclear. However, the PER2S478A mutant mouse line, which thus lacks the S478 phosphodegron, shows only a 30 to 60 min increase in the free-running behavioural period, and there is no clear period difference in embryonic fibroblasts derived from these mice (Masuda et al., 2020). This suggests a significant compensatory role for this additional CK1-dependent phosphodegron in regulating PER2 stability, either in the absence of, or in addition to, the S478 phosphodegron. PER2 is also ubiquitylated and targeted for degradation by the E3 ligase mouse double minute 2 homolog (MDM2) in a phosphorylation-independent manner, demonstrating that CK1-independent mechanisms also influence PER stability (Liu et al., 2018).
Complementary to its role in period determination, CK1 has been suggested as a means by which mammalian circadian rhythms are temperature compensated, meaning that they retain their ∼24-h period across the biologically relevant temperature range, giving them a temperature coefficient (Q10) close to 1. The activity of wild-type CK1δ and/or CK1ε on both the FASP and phosphodegron regions of PER2 is relatively temperature insensitive, with the balance of phosphorylation between the two sites relatively invariant across temperatures (Isojima et al., 2009). However, amino acid substitutions that influence the substrate preference of CK1δ and/or CK1ε for these two sites (Philpott et al., 2020) can alter temperature sensitivity of this kinase activity, causing cellular rhythms to become temperature sensitive (Shinohara et al., 2017). Recent work mapped these effects to the dynamics of α-helices E and F and their connecting loop in CK1δ. For example, CK1δ with a K224D mutation within the αF-helix retains normal activity at the non-consensus FASP priming site but displays reduced activity at the downstream phosphorylation sites (Philpott et al., 2020). This results in a significant increase in the Q10 compared to wild-type protein, with other mutations predicted to alter the dynamics of this helix showing similar effects (Shinohara et al., 2017). Similarly the tau mutation (R178C), which is situated adjacent to K224, leads to reduced activity at both the consensus and non-consensus FASP sites of PER2, but increased activity at its phosphodegron, thus exhibiting period shortening and reduced temperature compensation (Meng et al., 2008; Philpott et al., 2020; Shinohara et al., 2017).
Although classically considered to be the primary regulator of PER proteins, CK1 has also recently been shown to phosphorylate CLOCK, specifically within the repressive complex (Aryal et al., 2017). The exact site(s) and consequence of this phosphorylation is unknown, but it may contribute to the removal of CLOCK–BMAL1 complexes from their DNA target sequences at the beginning of the repressive phase. This would be analogous to the situation in Drosophila, where CLOCK only dissociates from DNA following phosphorylation by Doubletime (the Drosophila CK1δ/ε homolog), which is brought to the repressive complex by its association with PER (Kim et al., 2007). CK1 phosphorylation of mammalian CLOCK could occur at three sites within CLOCK that have recently been identified to be rhythmically phosphorylated in mouse liver, peaking at the time of maximal CLOCK binding to its target promoters (Robles et al., 2017). The comprehensive phosphoproteomic screen performed in this study not only confirmed some of the known phosphorylation sites within the core clock proteins, but also identified novel ones, such as S23 in Rev-Erbα. This, and other recent circadian proteomics work, provides a valuable resource in aiding our understanding of the regulation of circadian proteins by phosphorylation (Robles et al., 2017; Wang et al., 2018).
Casein kinase 2 (CK2) has also been shown to influence mammalian timekeeping; RNA knockdown or pharmacological inhibition of CK2α, and CK2β, increases circadian period, whereas overexpression shortens it (Maier et al., 2009; Tsuchiya et al., 2009; Oshima et al., 2019). CK2 purportedly stabilises PER2 through phosphorylation of its extreme N-terminus (Maier et al., 2009), although other work proposes that CK2 phosphorylation at S53 promotes PER2 degradation (Tsuchiya et al., 2009) (Fig. 2). More recent mass spectrometry analysis, however, indicates that the most significant site of CK2 phosphorylation on PER2 is S693, highlighting the need for further work to elucidate the activity of CK2 on PER2 (Oshima et al., 2019). CK2α also phosphorylates BMAL1 at S90, promoting BMAL1 nuclear entry; alanine substitution at this site increases the proportion of cytoplasmic BMAL1, preventing it from carrying out its transcriptional role (Tamaru et al., 2009). This phosphorylation is rhythmic due to rhythmic CRY association with BMAL1, with CRY bringing the less active CK2β subunit to the complex, thereby suppressing CK2α activity at this site (Tamaru et al., 2015).
Regulation by other kinases
A number of other kinases also play significant roles in regulating core clock proteins. GSK3β, another evolutionarily conserved kinase, phosphorylates a number of core clock components, including CRY2, BMAL1 and CLOCK, promoting their proteosomal degradation (Kurabayashi et al., 2010; Sahar et al., 2010; Spengler et al., 2009). Conversely, GSK3β-mediated phosphorylation of Rev-Erbα has a stabilising effect (Yin et al., 2006). In addition, GSK3β-induced phosphorylation of PER2, at a site distinct from that targeted by CK1, promotes nuclear translocation of PER2 (Iitaka et al., 2005). Strikingly, pharmacological inhibition of GSK3β shortens the circadian period, highlighting the complex and crucial role of this kinase in circadian timekeeping (Hirota et al., 2008). Given this, and the involvement of GSK3β in other circadian systems as diverse as Drosophila and the bread mould Neurospora crassa (Martinek et al., 2001; Tataroğlu et al., 2012), identifying the specific activities of GSK3β that regulate circadian rhythmicity could potentially provide significant insight into mammalian timekeeping.
Although classically considered a transcription factor, specific phosphorylation of BMAL1 allows it to directly regulate translation independently of its transcriptional role (Lipton et al., 2015). Here, rhythmic phosphorylation of BMAL1 residue S42 by the mTOR effector kinase S6K1 enables it to interact with cap-binding complexes, resulting in increased translation. This produces rhythmic total cellular protein synthesis, with puromycin incorporation (a measure of nascent protein production) varying by ∼15%, which is not observed in the absence of BMAL1 (Lipton et al., 2015).
Although comparatively little is known about post-translational regulation of ROR proteins, particularly within a circadian context, the ubiquitously expressed RORα4 isoform is negatively regulated by phosphorylation at T128 by extracellular signal-related kinase 2 (ERK2) (Lechtken et al., 2007). Alanine substitution at this site substantially increased the amount of Rev-Erbα required to attenuate RORα4 transcriptional activity at ROR response elements. Future work could determine how this regulatory mechanism plays a role in circadian timekeeping.
Cell cycle-dependent kinase 1 (CDK1) has been shown to phosphorylate Rev-Erbα at T275, prompting recognition by the FBXW7 E3 ubiquitin ligase complex, and subsequent degradation (Zhao et al., 2016), highlighting links between circadian rhythmicity and the cell cycle (Gaucher et al., 2018). The more ubiquitously expressed CDK5 also regulates circadian timekeeping through two mechanisms. Firstly, CDK5 phosphorylates PER2 at S394 in the PAS-B domain, modestly increasing its stability and facilitating its nuclear entry in complex with CRY1 (Brenna et al., 2019). Secondly, phosphorylation of CLOCK at T451 and T461 by CDK5 enhances CLOCK stability and transcriptional activity, leading to increased amplitude of expression of its targets (Kwak et al., 2013). CLOCK is phosphorylated at S845 by protein kinase B (AKT1) following insulin signalling, promoting its nuclear localisation (Luciano et al., 2018); S845A mutation in mice did not influence period, but did reduce expression of E-box-driven core clock protein transcripts (Per1, Per2 and Rev-Erbα) in some tissues.
The two mammalian CRY isoforms comprise a highly similar photolyase-homology region (PHR) with a primary FAD-binding pocket, a C-terminal coiled-coil-like (CC) helix and an intrinsically disordered C-terminal ‘tail’ (Czarna et al., 2013; Hirano et al., 2017; Nangle et al., 2013) (Fig. 3). Conservation between the PHRs of CRY1 and CRY2 suggests that findings regarding the PHR of CRY1, as discussed in the following paragraphs, likely also apply to CRY2, and vice versa. However, the proteins diverge in the identity of their C-terminal tails, providing a means for their differential regulation. Indeed, CRY2 is sequentially phosphorylated at S557 and S553 by dual-specificity tyrosine-phosphorylated and regulated kinase 1A (DYRK1A) and GSK3β, resulting in CRY2-specific degradation (Kurabayashi et al., 2010; Hirano et al., 2014). Conversely, phosphorylation of CRY1 at S588 stabilises it by antagonising the effect of the E3 ubiquitin ligase FBXL3, resulting in a longer circadian period (Papp et al., 2015); no equivalent stabilising activity has been identified in CRY2.
AMP-activated protein kinase (AMPK) also phosphorylates CRY1 at S71 and S280, reducing CRY1 stability and promoting its degradation by FBXL3 (Lamia et al., 2009; Siepka et al., 2007; Xing et al., 2013). FBXL21 also ubiquitylates CRY1, forming a more stable complex with slower activity in generating polyubiquitylated CRY1 than the FBXL3-CRY1 complex; thus, FBXL21 is thought to competitively inhibit the activity of FBXL3 towards CRY1, resulting in its stabilisation (Hirano et al., 2013; Yoo et al., 2013).
Recent work has identified ten phosphosites in CRY1, including those at S71 and S280, for which mutation to the phosphomimetic aspartate produced period effects or arrhythmicity in cells (Liu and Zhang, 2016) (Fig. 3). In a Herculean set of experiments, a subsequent study focused on a subset of these serine phosphosites that are situated on, or shortly after, the so-called phosphate-loop or ‘p-loop’ of CRY1 (Ode et al., 2017). This loop is adjacent to the FAD-binding pocket that modulates binding to co-factors, including FBXL3 (Xing et al., 2013). Aspartate substitution at these p-loop sites shortened cellular period and many, although not all, of these substitutions also shortened the circadian period in mouse models, with the period effect of these mutations being additive (Ode et al., 2017). The authors thus propose that, unlike the FASP region of PER2, phosphorylation of the CRY1 p-loop does not appear to be obligately sequential, but instead may act as a cumulative timer, where the order of phosphorylation of sites can vary, and it is instead the total number of phosphorylated sites that has an impact on cellular periodicity.
Owing to difficulties determining phosphatase specificity, few phosphatases have been identified to act selectively on mammalian clock proteins. Protein phosphatase 1 (PP1) acts antagonistically at sites in PER2 targeted by CK1ε to stabilise the protein, with expression of a dominant-negative PP1 accelerating PER2 degradation (Gallego et al., 2006; Lee et al., 2011). Additionally, PP5 increases CK1ε activity in vitro by removing autoinhibitory phosphorylation on the kinase; downregulation of PP5 results in decreased amplitude of PER expression and a potentially increased circadian period in fibroblasts (Partch et al., 2006). PP5 activity is inhibited by CRY in vitro (Partch et al., 2006), suggesting one way in which a clock protein could alter post-translational modification of its partner.
Moving beyond phosphorylation – other post-translational modifications
There is growing interest in the regulation of circadian rhythmicity by other post-translational modifications. BMAL1 is acetylated at K538 by the histone acetyltransferase TIP60 (also known as KAT5), which promotes elongation of the transcriptional targets of CLOCK–BMAL1 through the recruitment of elongation factors to form the BRD4–P-TEFb complex, leading to Pol II pause release (Petkau et al., 2019). It has been further proposed that phosphorylation of BMAL1 at S90 (discussed above) may be permissive for this acetylation (Tamaru et al., 2015). BMAL1 is also SUMOylated at K259 in a CLOCK-dependent manner, enhancing CLOCK–BMAL1 transcriptional activity, while simultaneously accelerating the ubiquitin-dependent proteolysis of BMAL1, which requires its SUMOylation (Cardone et al., 2005; Lee et al., 2008) (Fig. 3). Similarly, SUMOylation of RORα at K240 is also proposed to positively regulate its transcriptional activity (Hwang et al., 2009). This effect of SUMOylation is unusual, as SUMOylation is typically associated with reduced transcription of target genes (Rosonina et al., 2017). Further investigation of the mechanistic outcome of BMAL1 and RORα SUMOylation is therefore of particular interest.
Intriguingly, a number of serine residues within PER2, including those in the FASP region, are also capable of being O-GlcNAcylated (Kaasik et al., 2013). O-GlcNAcylation competes with CK1-mediated phosphorylation at these sites, and thus has been suggested as a point of integration of metabolic cues into the circadian clockwork. O-GlcNAc transferase (OGT) activity is regulated by GSK3β, demonstrating an additional role for circadian regulation by this enzyme (Kaasik et al., 2013) (Fig. 2). PER2 is also acetylated, with recent work suggesting that acetylation at K680 inhibits phosphorylation at the FASP priming site at S659 (mice) or S662 (humans), thereby influencing PER2 stability, although the relevant acetyl transferase, remains unknown (Levine et al., 2020). Deacetylation of PER2 by SIRT1 is important for turnover of the protein (Levine et al., 2020; Asher et al., 2008).
As described above, many post-translational modifications of core clock proteins regulate their stability. It is thus intuitive to posit that half-life of these proteins, particularly the CRY and PER proteins, might be a crucial determinant of circadian period. There is certainly a trend to this effect; manipulations that accelerate degradation of these proteins often lead to period shortening and vice versa (Meng et al., 2008; Ode et al., 2017). It is worth noting, therefore, that some mutations in and around the CRY1 p-loop that increase circadian period in cells do so without increasing protein half-life (Ode et al., 2017). This demonstrates that it is not merely the abundance of core clock proteins that is important for timekeeping, but also their molecular state. Currently, the determinants of such a molecular state, be they post-translational modifications, protein dynamics or association with other proteins, remain unknown, but this is certainly a fruitful topic for future research.
Perspectives
An expansive range of non-transcriptional elements regulate the core clock proteins. Indeed, the level of regulation that has come to light in the past decade has led to the postulation that post-transcriptional mechanisms, particularly post-translational modifications, might constitute a second, possibly more evolutionarily conserved, oscillator that acts in combination with the canonical TTFL, analogous to the prokaryotic cyanobacterial system (Ode and Ueda, 2018; Wong and O'Neill, 2018). Whether or not future research shows this to be the case, a greater and subtler knowledge of how clock proteins are regulated will substantially aid our understanding of mammalian timekeeping. Evidently, the findings outlined here are incomplete; the majority of our understanding pertaining to the regulation of the PER proteins has been elucidated for PER2, with little evidence to describe whether the same mechanisms also regulate PER1, or the much-neglected PER3, to name but one example. Some headway is being made towards this pursuit for the two CRY proteins and the CK1 isoforms CK1δ and CK1ε, which has produced some intriguing and insightful results showing strikingly different regulation and activity of these different proteins, which had previously been considered to be broadly identical in function (Fustin et al., 2018; Philpott et al., 2020; Rosensweig et al., 2018). Similarly, many unknowns still exist with regard to the regulation of the translation of core clock proteins, while attempts to visualise the protein complexes thought to make up the mammalian timekeeping machinery are still in early days (Aryal et al., 2017). Certainly, the observations discussed here represent only a small fraction of the myriad likely mechanisms of clock protein regulation, and we eagerly anticipate future progress in this direction.
Acknowledgements
We would like to thank Ryan Hamnett for patient and constructive comments, and members of the Partch Lab for discussion.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
P.C. is funded by the European Molecular Biology Organization (EMBO) grant ALTF 57-2019. C.L.P. is funded by National Institutes of Health grants GM107069 and GM121507. Deposited in PMC for release after 12 months.
References
- Anafi R. C., Francey L. J., Hogenesch J. B. and Kim J. (2017). CYCLOPS reveals human transcriptional rhythms in health and disease. Proc. Natl. Acad. Sci. USA 114, 5312-5317. 10.1073/pnas.1619320114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryal R. P., Kwak P. B., Tamayo A. G., Gebert M., Chiu P.-L., Walz T. and Weitz C. J. (2017). Macromolecular assemblies of the mammalian circadian clock. Mol. Cell 67, 770-782.e6. 10.1016/j.molcel.2017.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G., Gatfield D., Stratmann M., Reinke H., Dibner C., Kreppel F., Mostoslavsky R., Alt F. W. and Schibler U. (2008). SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134, 317-328. 10.1016/j.cell.2008.06.050 [DOI] [PubMed] [Google Scholar]
- Atger F., Gobet C., Marquis J., Martin E., Wang J., Weger B., Lefebvre G., Descombes P., Naef F. and Gachon F. (2015). Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proc. Natl. Acad. Sci. USA 112, E6579-E6588. 10.1073/pnas.1515308112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre A., Brown S. A., Marcacci L., Tronche F., Kellendonk C., Reichardt H. M., Schutz G. and Schibler U. (2000). Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289, 2344-2347. 10.1126/science.289.5488.2344 [DOI] [PubMed] [Google Scholar]
- Brenna A., Olejniczak I., Chavan R., Ripperger J. A., Langmesser S., Cameroni E., Hu Z., de Virgilio C., Dengjel J. and Albrecht U. (2019). Cyclin-dependent kinase 5 (CDK5) regulates the circadian clock. eLife 8, e50925 10.7554/eLife.50925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R., Li A., Cho H. Y., Lee B. and Obrietan K. (2010). Mammalian target of rapamycin signaling modulates photic entrainment of the suprachiasmatic circadian clock. J. Neurosci. 30, 6302-6314. 10.1523/JNEUROSCI.5482-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R., Gkogkas C. G., DE Zavalia N., Blum I. D., Yanagiya A., Tsukumo Y., Xu H., Lee C., Storch K.-F., Liu A. C. et al. (2015). Light-regulated translational control of circadian behavior by eIF4E phosphorylation. Nat. Neurosci. 18, 855-862. 10.1038/nn.4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone L., Hirayama J., Giordano F., Tamaru T., Palvimo J. J. and Sassone-Corsi P. (2005). Circadian clock control by SUMOylation of BMAL1. Science 309, 1390-1394. 10.1126/science.1110689 [DOI] [PubMed] [Google Scholar]
- Castelo-Szekely V., de Matos M., Tusup M., Pascolo S., Ule J. and Gatfield D. (2019). Charting DENR-dependent translation reinitiation uncovers predictive uORF features and links to circadian timekeeping via Clock. Nucleic Acids Res. 47, 5193-5209. 10.1093/nar/gkz261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Schirmer A., Lee Y., Lee H., Kumar V., Yoo S.-H., Takahashi J. S. and Lee C. (2009). Rhythmic PER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Mol. Cell 36, 417-430. 10.1016/j.molcel.2009.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., D'Alessandro M. and Lee C. (2013). miRNAs are required for generating a time delay critical for the circadian oscillator. Curr. Biol. 23, 1959-1968. 10.1016/j.cub.2013.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby P., Hamnett R., Putker M., Hoyle N. P., Reed M., Karam C. J., Maywood E. S., Stangherlin A., Chesham J. E., Hayter E. A. et al. (2019). Insulin/IGF-1 drives PERIOD synthesis to entrain circadian rhythms with feeding time. Cell 177, 896-909.e20. 10.1016/j.cell.2019.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crumbley C. and Burris T. P. (2011). Direct regulation of CLOCK expression by REV-ERB. PLoS ONE 6, e17290 10.1371/journal.pone.0017290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta M., Boudreau P., Dubeau-Laramée G., Cermakian N. and Boivin D. B. (2016). Simulated night shift disrupts circadian rhythms of immune functions in humans. J. Immunol. 196, 2466-2475. 10.4049/jimmunol.1502422 [DOI] [PubMed] [Google Scholar]
- Curtis A. M., Fagundes C. T., Yang G., Palsson-Mcdermott E. M., Wochal P., Mcgettrick A. F., Foley N. H., Early J. O., Chen L., Zhang H. et al. (2015). Circadian control of innate immunity in macrophages by miR-155 targeting Bmal1. Proc. Natl. Acad. Sci. USA 112, 7231-7236. 10.1073/pnas.1501327112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarna A., Berndt A., Singh H. R., Grudziecki A., Ladurner A. G., Timinszky G., Kramer A. and Wolf E. (2013). Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function. Cell 153, 1394-1405. 10.1016/j.cell.2013.05.011 [DOI] [PubMed] [Google Scholar]
- Dahlberg C. L., Nguyen E. Z., Goodlett D. and Kimelman D. (2009). Interactions between Casein kinase Iε (CKIε) and two substrates from disparate signaling pathways reveal mechanisms for substrate-kinase specificity. PLoS ONE 4, e4766 10.1371/journal.pone.0004766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decoursey P. J., Walker J. K. and Smith S. A. (2000). A circadian pacemaker in free-living chipmunks: essential for survival? J. Comp. Physiol. A 186, 169-180. 10.1007/s003590050017 [DOI] [PubMed] [Google Scholar]
- Dunlap J. C. (1999). Molecular bases for circadian clocks. Cell 96, 271-290. 10.1016/S0092-8674(00)80566-8 [DOI] [PubMed] [Google Scholar]
- Eide E. J., Woolf M. F., Kang H., Woolf P., Hurst W., Camacho F., Vielhaber E. L., Giovanni A. and Virshup D. M. (2005). Control of mammalian circadian rhythm by CKIε-regulated proteasome-mediated PER2 degradation. Mol. Cell. Biol. 25, 2795-2807. 10.1128/MCB.25.7.2795-2807.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng G. W. L., Edison, and Virshup D. M. (2017). Site-specific phosphorylation of casein kinase 1 delta (CK1delta) regulates its activity towards the circadian regulator PER2. PLoS One 12, e0177834 10.1371/journal.pone.0177834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian M. R., Sonenberg N. and Filipowicz W. (2010). Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79, 351-379. 10.1146/annurev-biochem-060308-103103 [DOI] [PubMed] [Google Scholar]
- Feeney K. A., Hansen L. L., Putker M., Olivares-Yañez C., Day J., Eades L. J., Larrondo L. F., Hoyle N. P., O'Neill J. S. and van Ooijen G. (2016). Daily magnesium fluxes regulate cellular timekeeping and energy balance. Nature 532, 375-379. 10.1038/nature17407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fustin J.-M., Doi M., Yamaguchi Y., Hida H., Nishimura S., Yoshida M., Isagawa T., Morioka M. S., Kakeya H., Manabe I. et al. (2013). RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155, 793-806. 10.1016/j.cell.2013.10.026 [DOI] [PubMed] [Google Scholar]
- Fustin J.-M., Kojima R., Itoh K., Chang H.-Y., Ye S., Zhuang B., Oji A., Gibo S., Narasimamurthy R., Virshup D. et al. (2018). Two Ck1δ transcripts regulated by m6A methylation code for two antagonistic kinases in the control of the circadian clock. Proc. Natl. Acad. Sci. USA 115, 5980-5985. 10.1073/pnas.1721371115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego M., Kang H. and Virshup D. M. (2006). Protein phosphatase 1 regulates the stability of the circadian protein PER2. Biochem. J. 399, 169-175. 10.1042/BJ20060678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mauriño S. M., Rivero-Rodríguez F., Velázquez-Cruz A., Hernández-Vellisca M., Díaz-Quintana A., De la Rosa M. A. and Díaz-Moreno I. (2017). RNA binding protein regulation and cross-talk in the control of AU-rich mRNA fate. Front. Mol. Biosci. 4, 71 10.3389/fmolb.2017.00071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher J., Montellier E. and Sassone-Corsi P. (2018). Molecular cogs: interplay between circadian clock and cell cycle. Trends Cell Biol. 28, 368-379. 10.1016/j.tcb.2018.01.006 [DOI] [PubMed] [Google Scholar]
- Giamas G., Hirner H., Shoshiashvili L., Grothey A., Gessert S., Kühl M., Henne-Bruns D., Vorgias C. E. and Knippschild U. (2007). Phosphorylation of CK1δ: identification of Ser370 as the major phosphorylation site targeted by PKA in vitro and in vivo. Biochem. J. 406, 389-398. 10.1042/BJ20070091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano A., Yumimoto K., Tsunematsu R., Matsumoto M., Oyama M., Kozuka-Hata H., Nakagawa T., Lanjakornsiripan D., Nakayama K. I. and Fukada Y. (2013). FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell 152, 1106-1118. 10.1016/j.cell.2013.01.054 [DOI] [PubMed] [Google Scholar]
- Hirano A., Kurabayashi N., Nakagawa T., Shioi G., Todo T., Hirota T. and Fukada Y. (2014). In vivo role of phosphorylation of cryptochrome 2 in the mouse circadian clock. Mol. Cell. Biol. 34, 4464-4473. 10.1128/MCB.00711-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano A., Braas D., Fu Y.-H. and Ptáček L. J. (2017). FAD regulates CRYPTOCHROME protein stability and circadian clock in mice. Cell Rep. 19, 255-266. 10.1016/j.celrep.2017.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T., Lewis W. G., Liu A. C., Lee J. W., Schultz P. G. and Kay S. A. (2008). A chemical biology approach reveals period shortening of the mammalian circadian clock by specific inhibition of GSK-3beta. Proc. Natl. Acad. Sci. USA 105, 20746-20751. 10.1073/pnas.0811410106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyle N. P., Seinkmane E., Putker M., Feeney K. A., Krogager T. P., Chesham J. E., Bray L. K., Thomas J. M., Dunn K., Blaikley J. et al. (2017). Circadian actin dynamics drive rhythmic fibroblast mobilization during wound healing. Sci. Transl. Med. 9, eaal2774 10.1126/scitranslmed.aal2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang E. J., Lee J. M., Jeong J., Park J. H., Yang Y., Lim J.-S., Kim J. H., Baek S. H. and Kim K. I. (2009). SUMOylation of RORα potentiates transcriptional activation function. Biochem. Biophys. Res. Commun. 378, 513-517. 10.1016/j.bbrc.2008.11.072 [DOI] [PubMed] [Google Scholar]
- Iitaka C., Miyazaki K., Akaike T. and Ishida N. (2005). A role for glycogen synthase kinase-3β in the mammalian circadian clock. J. Biol. Chem. 280, 29397-29402. 10.1074/jbc.M503526200 [DOI] [PubMed] [Google Scholar]
- Ikeda R., Tsuchiya Y., Koike N., Umemura Y., Inokawa H., Ono R., Inoue M., Sasawaki Y., Grieten T., Okubo N. et al. (2019). REV-ERBα and REV-ERBβ function as key factors regulating Mammalian Circadian Output. Sci. Rep. 9, 10171 10.1038/s41598-019-46656-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isojima Y., Nakajima M., Ukai H., Fujishima H., Yamada R. G., Masumoto K.-H., Kiuchi R., Ishida M., Ukai-Tadenuma M., Minami Y. et al. (2009). CKIε/δ-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc. Natl. Acad. Sci. USA 106, 15744-15749. 10.1073/pnas.0908733106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang C., Lahens N. F., Hogenesch J. B. and Sehgal A. (2015). Ribosome profiling reveals an important role for translational control in circadian gene expression. Genome Res. 25, 1836-1847. 10.1101/gr.191296.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janich P., Arpat A. B., Castelo-Szekely V., Lopes M. and Gatfield D. (2015). Ribosome profiling reveals the rhythmic liver translatome and circadian clock regulation by upstream open reading frames. Genome Res. 25, 1848-1859. 10.1101/gr.195404.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouffe C., Cretenet G., Symul L., Martin E., Atger F., Naef F. and Gachon F. (2013). The circadian clock coordinates ribosome biogenesis. PLoS Biol. 11, e1001455 10.1371/journal.pbio.1001455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y., Ryu H. G., Kim S. W., Lee K.-H., Gu S., Yi H., Ku H.-O., Jang S. K. and Kim K.-T. (2019). The RNA-binding protein hnRNP Q represses translation of the clock gene Bmal1 in murine cells. J. Biol. Chem. 294, 7682-7691. 10.1074/jbc.RA118.006947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaasik K., Kivimäe S., Allen J. J., Chalkley R. J., Huang Y., Baer K., Kissel H., Burlingame A. L., Shokat K. M., Ptáček L. J. et al. (2013). Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab. 17, 291-302. 10.1016/j.cmet.2012.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. Y., Ko H. W., Yu W., Hardin P. E. and Edery I. (2007). A DOUBLETIME kinase binding domain on the Drosophila PERIOD protein is essential for its hyperphosphorylation, transcriptional repression, and circadian clock function. Mol. Cell. Biol. 27, 5014-5028. 10.1128/MCB.02339-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.-Y., Woo K.-C., Lee K.-H., Kim T.-D. and Kim K.-T. (2010). hnRNP Q and PTB modulate the circadian oscillation of mouse Rev-erb α via IRES-mediated translation. Nucleic Acids Res. 38, 7068-7078. 10.1093/nar/gkq569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.-Y., Kwak E., Kim S.-H., Lee K.-H., Woo K.-C. and Kim K.-T. (2011). hnRNP Q mediates a phase-dependent translation-coupled mRNA decay of mouse Period3. Nucleic Acids Res. 39, 8901-8914. 10.1093/nar/gkr605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-H., Lee K.-H., Kim D.-Y., Kwak E., Kim S. and Kim K.-T. (2015). Rhythmic control of mRNA stability modulates circadian amplitude of mouse Period3 mRNA. J. Neurochem. 132, 642-656. 10.1111/jnc.13027 [DOI] [PubMed] [Google Scholar]
- Koike N., Yoo S.-H., Huang H.-C., Kumar V., Lee C., Kim T.-K. and Takahashi J. S. (2012). Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349-354. 10.1126/science.1226339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S., Matsumoto K., Hirose M., Shimada M., Nagano M., Shigeyoshi Y., Hoshino S., Ui-Tei K., Saigo K., Green C. B. et al. (2007). LARK activates posttranscriptional expression of an essential mammalian clock protein, PERIOD1. Proc. Natl. Acad. Sci. USA 104, 1859-1864. 10.1073/pnas.0607567104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurabayashi N., Hirota T., Sakai M., Sanada K. and Fukada Y. (2010). DYRK1A and glycogen synthase kinase 3β, a dual-kinase mechanism directing proteasomal degradation of CRY2 for circadian timekeeping. Mol. Cell. Biol. 30, 1757-1768. 10.1128/MCB.01047-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak E., Kim T.-D. and Kim K.-T. (2006). Essential role of 3′-untranslated region-mediated mRNA decay in circadian oscillations of mouse Period3 mRNA. J. Biol. Chem. 281, 19100-19106. 10.1074/jbc.M511927200 [DOI] [PubMed] [Google Scholar]
- Kwak Y., Jeong J., Lee S., Park Y.-U., Lee S.-A., Han D.-H., Kim J.-H., Ohshima T., Mikoshiba K., Suh Y.-H. et al. (2013). Cyclin-dependent kinase 5 (Cdk5) regulates the function of CLOCK protein by direct phosphorylation. J. Biol. Chem. 288, 36878-36889. 10.1074/jbc.M113.494856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia K. A., Sachdeva U. M., Ditacchio L., Williams E. C., Alvarez J. G., Egan D. F., Vasquez D. S., Juguilon H., Panda S., Shaw R. J. et al. (2009). AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 326, 437-440. 10.1126/science.1172156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtken A., Hörnig M., Werz O., Corvey N., Zündorf I., Dingermann T., Brandes R. and Steinhilber D. (2007). Extracellular signal-regulated kinase-2 phosphorylates RORα4 in vitro. Biochem. Biophys. Res. Commun. 358, 890-896. 10.1016/j.bbrc.2007.05.016 [DOI] [PubMed] [Google Scholar]
- Lee C., Etchegaray J.-P., Cagampang F. R. A., Loudon A. S. I. and Reppert S. M. (2001). Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107, 855-867. 10.1016/S0092-8674(01)00610-9 [DOI] [PubMed] [Google Scholar]
- Lee J., Lee Y., Lee M. J., Park E., Kang S. H., Chung C. H., Lee K. H. and Kim K. (2008). Dual modification of BMAL1 by SUMO2/3 and ubiquitin promotes circadian activation of the CLOCK/BMAL1 complex. Mol. Cell. Biol. 28, 6056-6065. 10.1128/MCB.00583-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.-M., Chen R., Kim H., Etchegaray J.-P., Weaver D. R. and Lee C. (2011). The period of the circadian oscillator is primarily determined by the balance between casein kinase 1 and protein phosphatase 1. Proc. Natl. Acad. Sci. USA 108, 16451-16456. 10.1073/pnas.1107178108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.-H., Kim S.-H., Kim D.-Y., Kim S. and Kim K.-T. (2012a). Internal ribosomal entry site-mediated translation is important for rhythmic PERIOD1 expression. PLoS ONE 7, e37936 10.1371/journal.pone.0037936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.-H., Woo K.-C., Kim D.-Y., Kim T.-D., Shin J., Park S. M., Jang S. K. and Kim K.-T. (2012b). Rhythmic interaction between Period1 mRNA and hnRNP Q leads to circadian time-dependent translation. Mol. Cell. Biol. 32, 717-728. 10.1128/MCB.06177-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.-H., Kim S.-H., Lee H.-R., Kim W., Kim D.-Y., Shin J.-C., Yoo S.-H. and Kim K.-T. (2013). MicroRNA-185 oscillation controls circadian amplitude of mouse Cryptochrome 1 via translational regulation. Mol. Biol. Cell 24, 2248-2255. 10.1091/mbc.e12-12-0849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.-H., Kim S.-H., Kim H.-J., Kim W., Lee H.-R., Jung Y., Choi J.-H., Hong K. Y., Jang S. K. and Kim K.-T. (2014). AUF1 contributes to Cryptochrome1 mRNA degradation and rhythmic translation. Nucleic Acids Res. 42, 3590-3606. 10.1093/nar/gkt1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppek K., Das R. and Barna M. (2018). Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 19, 158-174. 10.1038/nrm.2017.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine D. C., Hong H., Weidemann B. J., Ramsey K. M., Affinati A. H., Schmidt M. S., Cedernaes J., Omura C., Braun R., Lee C. et al. (2020). NAD(+) controls circadian reprogramming through PER2 nuclear translocation to counter aging. Mol. Cell 78, 835-849.e7. 10.1016/j.molcel.2020.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Wang Y., Wang F., Hu L.-F. and Liu C.-F. (2017). A new perspective for Parkinson's disease: circadian rhythm. Neurosci. Bull. 33, 62-72. 10.1007/s12264-016-0089-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim I., Jung Y., Kim D.-Y. and Kim K.-T. (2016). HnRNP Q has a suppressive role in the translation of mouse cryptochrome1. PLoS ONE 11, e0159018 10.1371/journal.pone.0159018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton J. O., Yuan E. D., Boyle L. M., Ebrahimi-Fakhari D., Kwiatkowski E., Nathan A., Güttler T., Davis F., Asara J. M. and Sahin M. (2015). The circadian protein BMAL1 regulates translation in response to S6K1-mediated phosphorylation. Cell 161, 1138-1151. 10.1016/j.cell.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N. and Zhang E. E. (2016). Phosphorylation regulating the ratio of intracellular CRY1 protein determines the circadian period. Front. Neurol. 7, 159 10.3389/fneur.2016.00159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A. C., Tran H. G., Zhang E. E., Priest A. A., Welsh D. K. and Kay S. A. (2008). Redundant function of REV-ERBα and β and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 4, e1000023 10.1371/journal.pgen.1000023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Zou X., Gotoh T., Brown A. M., Jiang L., Wisdom E. L., Kim J. K. and Finkielstein C. V. (2018). Distinct control of PERIOD2 degradation and circadian rhythms by the oncoprotein and ubiquitin ligase MDM2. Sci. Signal. 11 10.1126/scisignal.aau0715 [DOI] [PubMed] [Google Scholar]
- Lowrey P. L., Shimomura K., Antoch M. P., Yamazaki S., Zemenides P. D., Ralph M. R., Menaker M. and Takahashi J. S. (2000). Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 288, 483-492. 10.1126/science.288.5465.483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano A. K., Zhou W., Santana J. M., Kyriakides C., Velazquez H. and Sessa W. C. (2018). CLOCK phosphorylation by AKT regulates its nuclear accumulation and circadian gene expression in peripheral tissues. J. Biol. Chem. 293, 9126-9136. 10.1074/jbc.RA117.000773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lück S., Thurley K., Thaben P. F. and Westermark P. O. (2014). Rhythmic degradation explains and unifies circadian transcriptome and proteome data. Cell Rep. 9, 741-751. 10.1016/j.celrep.2014.09.021 [DOI] [PubMed] [Google Scholar]
- Maier B., Wendt S., Vanselow J. T., Wallach T., Reischl S., Oehmke S., Schlosser A. and Kramer A. (2009). A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev. 23, 708-718. 10.1101/gad.512209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinek S., Inonog S., Manoukian A. S. and Young M. W. (2001). A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell 105, 769-779. 10.1016/S0092-8674(01)00383-X [DOI] [PubMed] [Google Scholar]
- Masuda S., Narasimamurthy R., Yoshitane H., Kim J. K., Fukada Y. and Virshup D. M. (2020). Mutation of a PER2 phosphodegron perturbs the circadian phosphoswitch. Proc. Natl. Acad. Sci. USA 117, 10888-10896. 10.1073/pnas.2000266117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauvoisin D., Wang J., Jouffe C., Martin E., Atger F., Waridel P., Quadroni M., Gachon F. and Naef F. (2014). Circadian clock-dependent and -independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proc. Natl. Acad. Sci. USA 111, 167-172. 10.1073/pnas.1314066111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcglincy N. J., Valomon A., Chesham J. E., Maywood E. S., Hastings M. H. and Ule J. (2012). Regulation of alternative splicing by the circadian clock and food related cues. Genome Biol. 13, R54 10.1186/gb-2012-13-6-r54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet J. S., Rodriguez J., Abruzzi K. C. and Rosbash M. (2012). Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. eLife 1, e00011 10.7554/eLife.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q.-J., Logunova L., Maywood E. S., Gallego M., Lebiecki J., Brown T. M., Sládek M., Semikhodskii A. S., Glossop N. R. J., Piggins H. D. et al. (2008). Setting clock speed in mammals: the CK1ɛ tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron 58, 78-88. 10.1016/j.neuron.2008.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q.-J., Maywood E. S., Bechtold D. A., Lu W.-Q., Li J., Gibbs J. E., Dupre S. M., Chesham J. E., Rajamohan F., Knafels J. et al. (2010). Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. Proc. Natl. Acad. Sci. USA 107, 15240-15245. 10.1073/pnas.1005101107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyuhas O. and Kahan T. (2015). The race to decipher the top secrets of TOP mRNAs. Biochim. Biophys. Acta 1849, 801-811. 10.1016/j.bbagrm.2014.08.015 [DOI] [PubMed] [Google Scholar]
- Nagel R., Clijsters L. and Agami R. (2009). The miRNA-192/194 cluster regulates the Period gene family and the circadian clock. FEBS J. 276, 5447-5455. 10.1111/j.1742-4658.2009.07229.x [DOI] [PubMed] [Google Scholar]
- Nangle S., Xing W. and Zheng N. (2013). Crystal structure of mammalian cryptochrome in complex with a small molecule competitor of its ubiquitin ligase. Cell Res. 23, 1417-1419. 10.1038/cr.2013.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimamurthy R., Hunt S. R., Lu Y., Fustin J.-M., Okamura H., Partch C. L., Forger D. B., Kim J. K. and Virshup D. M. (2018). CK1δ/ε protein kinase primes the PER2 circadian phosphoswitch. Proc. Natl. Acad. Sci. USA 115, 5986-5991. 10.1073/pnas.1721076115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ode K. L. and Ueda H. R. (2018). Design principles of phosphorylation-dependent timekeeping in eukaryotic Circadian clocks. Cold Spring Harb. Perspect. Biol. 10, a028357 10.1101/cshperspect.a028357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ode K. L., Ukai H., Susaki E. A., Narumi R., Matsumoto K., Hara J., Koide N., Abe T., Kanemaki M. T., Kiyonari H. et al. (2017). Knockout-rescue embryonic stem cell-derived mouse reveals circadian-period control by quality and quantity of CRY1. Mol. Cell 65, 176-190. 10.1016/j.molcel.2016.11.022 [DOI] [PubMed] [Google Scholar]
- Ohsaki K., Oishi K., Kozono Y., Nakayama K., Nakayama K. I. and Ishida N. (2008). The role of β-TrCP1 and β-TrCP2 in circadian rhythm generation by mediating degradation of clock protein PER2. J. Biochem. 144, 609-618. 10.1093/jb/mvn112 [DOI] [PubMed] [Google Scholar]
- Oshima T., Niwa Y., Kuwata K., Srivastava A., Hyoda T., Tsuchiya Y., Kumagai M., Tsuyuguchi M., Tamaru T., Sugiyama A. et al. (2019). Cell-based screen identifies a new potent and highly selective CK2 inhibitor for modulation of circadian rhythms and cancer cell growth. Sci. Adv. 5, eaau9060 10.1126/sciadv.aau9060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp S. J., Huber A.-L., Jordan S. D., Kriebs A., Nguyen M., Moresco J. J., Yates J. R. III and Lamia K. A. (2015). DNA damage shifts circadian clock time via Hausp-dependent Cry1 stabilization. eLife 4, e04883 10.7554/eLife.04883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partch C. L. (2020). Orchestration of circadian timing by macromolecular protein assemblies. J. Mol. Biol. 432, 3426-3448. 10.1016/j.jmb.2019.12.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partch C. L., Shields K. F., Thompson C. L., Selby C. P. and Sancar A. (2006). Posttranslational regulation of the mammalian circadian clock by cryptochrome and protein phosphatase 5. Proc. Natl. Acad. Sci. USA 103, 10467-10472. 10.1073/pnas.0604138103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkau N., Budak H., Zhou X., Oster H. and Eichele G. (2019). Acetylation of BMAL1 by TIP60 controls BRD4-P-TEFb recruitment to circadian promoters. eLife 8, e43235 10.7554/eLife.43235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott J. M., Narasimamurthy R., Ricci C. G., Freeberg A. M., Hunt S. R., Yee L. E., Pelofsky R. S., Tripathi S., Virshup D. M. and Partch C. L. (2020). Casein kinase 1 dynamics underlie substrate selectivity and the PER2 circadian phosphoswitch. eLife 9, e52343 10.7554/eLife.52343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh C. S. (1960). Circadian rhythms and the circadian organization of living systems. Cold Spring Harb. Symp. Quant. Biol. 25, 159-184. 10.1101/SQB.1960.025.01.015 [DOI] [PubMed] [Google Scholar]
- Ralph M. R. and Menaker M. (1988). A mutation of the circadian system in golden hamsters. Science 241, 1225-1227. 10.1126/science.3413487 [DOI] [PubMed] [Google Scholar]
- Ramanathan C., Kathale N. D., Liu D., Lee C., Freeman D. A., Hogenesch J. B., Cao R. and Liu A. C. (2018). mTOR signaling regulates central and peripheral circadian clock function. PLoS Genet. 14, e1007369 10.1371/journal.pgen.1007369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A. B. and O'Neill J. S. (2010). Healthy clocks, healthy body, healthy mind. Trends Cell Biol. 20, 36-44. 10.1016/j.tcb.2009.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A. B., Karp N. A., Maywood E. S., Sage E. A., Deery M., O'Neill J. S., Wong G. K. Y., Chesham J., Odell M., Lilley K. S. et al. (2006). Circadian orchestration of the hepatic proteome. Curr. Biol. 16, 1107-1115. 10.1016/j.cub.2006.04.026 [DOI] [PubMed] [Google Scholar]
- Reischl S., Vanselow K., Westermark P. O., Thierfelder N., Maier B., Herzel H. and Kramer A. (2007). Beta-TrCP1-mediated degradation of PERIOD2 is essential for circadian dynamics. J. Biol. Rhythms 22, 375-386. 10.1177/0748730407303926 [DOI] [PubMed] [Google Scholar]
- Rey G., Cesbron F., Rougemont J., Reinke H., Brunner M. and Naef F. (2011). Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 9, e1000595 10.1371/journal.pbio.1000595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles M. S., Cox J. and Mann M. (2014). In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS Genet. 10, e1004047 10.1371/journal.pgen.1004047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles M. S., Humphrey S. J. and Mann M. (2017). Phosphorylation is a central mechanism for circadian control of metabolism and physiology. Cell Metab. 25, 118-127. 10.1016/j.cmet.2016.10.004 [DOI] [PubMed] [Google Scholar]
- Roenneberg T. and Merrow M. (2005). Circadian clocks - the fall and rise of physiology. Nat. Rev. Mol. Cell Biol. 6, 965-971. 10.1038/nrm1766 [DOI] [PubMed] [Google Scholar]
- Rosensweig C., Reynolds K. A., Gao P., Laothamatas I., Shan Y., Ranganathan R., Takahashi J. S. and Green C. B. (2018). An evolutionary hotspot defines functional differences between CRYPTOCHROMES. Nat. Commun. 9, 1138 10.1038/s41467-018-03503-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosonina E., Akhter A., Dou Y., Babu J. and SRI Theivakadadcham V. S. (2017). Regulation of transcription factors by sumoylation. Transcription 8, 220-231. 10.1080/21541264.2017.1311829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed Y. and Abbott S. M. (2017). Circadian disruption associated with Alzheimer's disease. Curr. Neurol. Neurosci. Rep. 17, 29 10.1007/s11910-017-0745-y [DOI] [PubMed] [Google Scholar]
- Sahar S., Zocchi L., Kinoshita C., Borrelli E. and Sassone-Corsi P. (2010). Regulation of BMAL1 protein stability and circadian function by GSK3β-mediated phosphorylation. PLoS ONE 5, e8561 10.1371/journal.pone.0008561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado-Delgado R. C., Saderi N., Basualdo M. C., Guerrero-Vargas N. N., Escobar C. and Buijs R. M. (2013). Shift work or food intake during the rest phase promotes metabolic disruption and desynchrony of liver genes in male rats. PLoS ONE 8, e60052 10.1371/journal.pone.0060052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer F. A. J. L., Hilton M. F., Mantzoros C. S. and Shea S. A. (2009). Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc. Natl. Acad. Sci. USA 106, 4453-4458. 10.1073/pnas.0808180106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleich S., Strassburger K., Janiesch P. C., Koledachkina T., Miller K. K., Haneke K., Cheng Y.-S., Küchler K., Stoecklin G., Duncan K. E. et al. (2014). DENR-MCT-1 promotes translation re-initiation downstream of uORFs to control tissue growth. Nature 512, 208-212. 10.1038/nature13401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanware N. P., Hutchinson J. A., Kim S. H., Zhan L., Bowler M. J. and Tibbetts R. S. (2011). Casein kinase 1-dependent phosphorylation of familial advanced sleep phase syndrome-associated residues controls PERIOD 2 stability. J. Biol. Chem. 286, 12766-12774. 10.1074/jbc.M111.224014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara Y., Koyama Y. M., Ukai-Tadenuma M., Hirokawa T., Kikuchi M., Yamada R. G., Ukai H., Fujishima H., Umehara T., Tainaka K. et al. (2017). Temperature-sensitive substrate and product binding underlie temperature-compensated phosphorylation in the clock. Mol. Cell 67, 783-798.e20. 10.1016/j.molcel.2017.08.009 [DOI] [PubMed] [Google Scholar]
- Shirogane T., Jin J., Ang X. L. and Harper J. W. (2005). SCFβ-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J. Biol. Chem. 280, 26863-26872. 10.1074/jbc.M502862200 [DOI] [PubMed] [Google Scholar]
- Siepka S. M., Yoo S.-H., Park J., Song W., Kumar V., Hu Y., Lee C. and Takahashi J. S. (2007). Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell 129, 1011-1023. 10.1016/j.cell.2007.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinturel F., Gerber A., Mauvoisin D., Wang J., Gatfield D., Stubblefield J. J., Green C. B., Gachon F. and Schibler U. (2017). Diurnal oscillations in liver mass and cell size accompany ribosome assembly cycles. Cell 169, 651-663.e14. 10.1016/j.cell.2017.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler M. L., Kuropatwinski K. K., Schumer M. and Antoch M. P. (2009). A serine cluster mediates BMAL1-dependent CLOCK phosphorylation and degradation. Cell Cycle 8, 4138-4146. 10.4161/cc.8.24.10273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi J. S. (2017). Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164-179. 10.1038/nrg.2016.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru T., Hirayama J., Isojima Y., Nagai K., Norioka S., Takamatsu K. and Sassone-Corsi P. (2009). CK2α phosphorylates BMAL1 to regulate the mammalian clock. Nat. Struct. Mol. Biol. 16, 446-448. 10.1038/nsmb.1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru T., Hattori M., Honda K., Nakahata Y., Sassone-Corsi P., van der Horst G. T. J., Ozawa T. and Takamatsu K. (2015). CRY drives cyclic CK2-mediated BMAL1 phosphorylation to control the mammalian circadian clock. PLoS Biol. 13, e1002293 10.1371/journal.pbio.1002293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tataroğlu O., Lauinger L., Sancar G., Jakob K., Brunner M. and Diernfellner A. C. R. (2012). Glycogen synthase kinase is a regulator of the circadian clock of Neurospora crassa. J. Biol. Chem. 287, 36936-36943. 10.1074/jbc.M112.396622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh K. L., Jones C. R., He Y., Eide E. J., Hinz W. A., Virshup D. M., Ptacek L. J. and Fu Y. H. (2001). An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 291, 1040-1043. 10.1126/science.1057499 [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y., Akashi M., Matsuda M., Goto K., Miyata Y., Node K. and Nishida E. (2009). Involvement of the protein kinase CK2 in the regulation of mammalian circadian rhythms. Sci. Signal. 2, ra26 10.1126/scisignal.2000305 [DOI] [PubMed] [Google Scholar]
- Um J. H., Yang S., Yamazaki S., Kang H., Viollet B., Foretz M. and Chung J. H. (2007). Activation of 5′-AMP-activated kinase with diabetes drug metformin induces casein kinase Iɛ (CKIɛ)-dependent degradation of clock protein mPer2. J. Biol. Chem. 282, 20794-20798. 10.1074/jbc.C700070200 [DOI] [PubMed] [Google Scholar]
- Vanselow K., Vanselow J. T., Westermark P. O., Reischl S., Maier B., Korte T., Herrmann A., Herzel H., Schlosser A. and Kramer A. (2006). Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS). Genes Dev. 20, 2660-2672. 10.1101/gad.397006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Song L., Liu M., Ge R., Zhou Q., Liu W., Li R., Qie J., Zhen B., Wang Y. et al. (2018). A proteomics landscape of circadian clock in mouse liver. Nat. Commun. 9, 1553 10.1038/s41467-018-03898-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh D. K., Imaizumi T. and Kay S. A. (2005). Real-time reporting of circadian-regulated gene expression by luciferase imaging in plants and mammalian cells. Methods Enzymol. 393, 269-288. 10.1016/S0076-6879(05)93011-5 [DOI] [PubMed] [Google Scholar]
- Woelfle M. A., Ouyang Y., Phanvijhitsiri K. and Johnson C. H. (2004). The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Curr. Biol. 14, 1481-1486. 10.1016/j.cub.2004.08.023 [DOI] [PubMed] [Google Scholar]
- Wong D. C. S. and O'Neill J. S. (2018). Non-transcriptional processes in circadian rhythm generation. Curr. Opin. Physiol. 5, 117-132. 10.1016/j.cophys.2018.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo K.-C., Kim T.-D., Lee K.-H., Kim D.-Y., Kim W., Lee K.-Y. and Kim K.-T. (2009). Mouse period 2 mRNA circadian oscillation is modulated by PTB-mediated rhythmic mRNA degradation. Nucleic Acids Res. 37, 26-37. 10.1093/nar/gkn893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo K.-C., Ha D.-C., Lee K.-H., Kim D.-Y., Kim T.-D. and Kim K.-T. (2010). Circadian amplitude of cryptochrome 1 is modulated by mRNA stability regulation via cytoplasmic hnRNP D oscillation. Mol. Cell. Biol. 30, 197-205. 10.1128/MCB.01154-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing W., Busino L., Hinds T. R., Marionni S. T., Saifee N. H., Bush M. F., Pagano M. and Zheng N. (2013). SCF(FBXL3) ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature 496, 64-68. 10.1038/nature11964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L., Wang J., Klein P. S. and Lazar M. A. (2006). Nuclear receptor Rev-erbα is a critical lithium-sensitive component of the circadian clock. Science 311, 1002-1005. 10.1126/science.1121613 [DOI] [PubMed] [Google Scholar]
- Yoo S.-H., Yamazaki S., Lowrey P. L., Shimomura K., Ko C. H., Buhr E. D., Siepka S. M., Hong H.-K., Oh W. J., Yoo O. J. et al. (2004). PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. USA 101, 5339-5346. 10.1073/pnas.0308709101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.-H., Mohawk J. A., Siepka S. M., Shan Y., Huh S. K., Hong H.-K., Kornblum I., Kumar V., Koike N., Xu M. et al. (2013). Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell 152, 1091-1105. 10.1016/j.cell.2013.01.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.-H., Kojima S., Shimomura K., Koike N., Buhr E. D., Furukawa T., Ko C. H., Gloston G., Ayoub C., Nohara K. et al. (2017). Period2 3'-UTR and microRNA-24 regulate circadian rhythms by repressing PERIOD2 protein accumulation. Proc. Natl. Acad. Sci. USA 114, E8855-E8864. 10.1073/pnas.1706611114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Lahens N. F., Ballance H. I., Hughes M. E. and Hogenesch J. B. (2014). A circadian gene expression atlas in mammals: implications for biology and medicine. Proc. Natl. Acad. Sci. USA 111, 16219-16224. 10.1073/pnas.1408886111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Hirota T., Han X., Cho H., Chong L.-W., Lamia K., Liu S., Atkins A. R., Banayo E., Liddle C. et al. (2016). Circadian Amplitude Regulation via FBXW7-Targeted REV-ERBα Degradation. Cell 165, 1644-1657. 10.1016/j.cell.2016.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B. S., Roundtree I. A. and He C. (2017). Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 18, 31-42. 10.1038/nrm.2016.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Kim J. K., Eng G. W. L., Forger D. B. and Virshup D. M. (2015). A Period2 phosphoswitch regulates and temperature compensates circadian period. Mol. Cell 60, 77-88. 10.1016/j.molcel.2015.08.022 [DOI] [PubMed] [Google Scholar]