Figure 1.
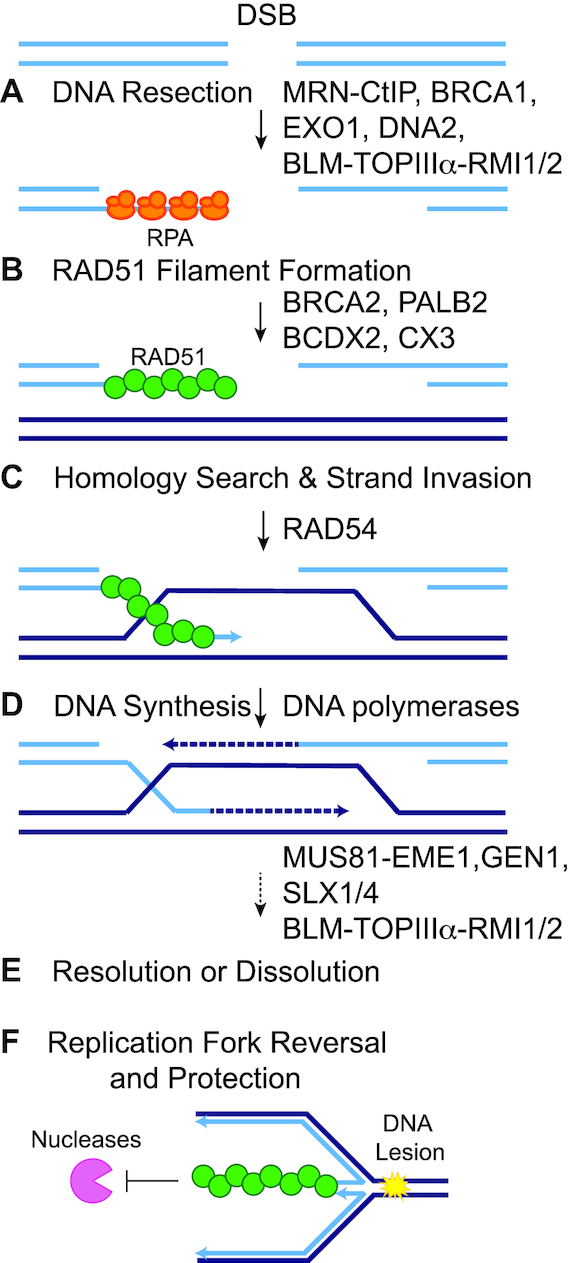
Schematic of RAD51 function during HR and replication fork reversal and protection. After a DSB occurs, the cell can use the HR pathway to repair the break using a homologous template (dark blue lines). (A) The DNA ends are resected to form 3′ ssDNA overhangs that are coated with RPA (orange ovals). Short-range DNA end resection is mediated by MRE11–RAD50–NBS1 (MRN) with CtIP and long-range DNA end resection is mediated by EXO1 or BLM–TOPIIIα–RMI1/2 with DNA2. BRCA1 also has an important function during DNA end resection. (B) RPA is then displaced by RAD51 (green circles), which subsequently forms a nucleoprotein filament. RAD51 filament formation is aided by PALB2, BRCA2 and the RAD51 paralog sub-complexes (BCDX2 and CX3). (C) The RAD51 nucleoprotein filament invades the homologous template in search for a homologous sequence. The strand invasion by the RAD51 filament forms a D-loop structure. RAD54 aids in these processes. (D) RAD51 is displaced and the DNA is extended by polymerases that copy the missing nucleotides from the repair template. (E) The second end of the DSB is captured and the DNA intermediate is resolved through resolution or dissolution, resulting in either a crossover or non-crossover product. HR resolution is aided by MUS81–EME1, GEN1 or SLX1/4, whereas dissolution occurs through the action of BLM–TOPIIIα–RMI1/2. (F) RAD51 functions at stalled replication forks. When the replication fork encounters a fork-blocking lesion (yellow starburst), RAD51 promotes replication fork reversal and protects the nascent strands of DNA from degradation by exonucleases (pink pac-man).
