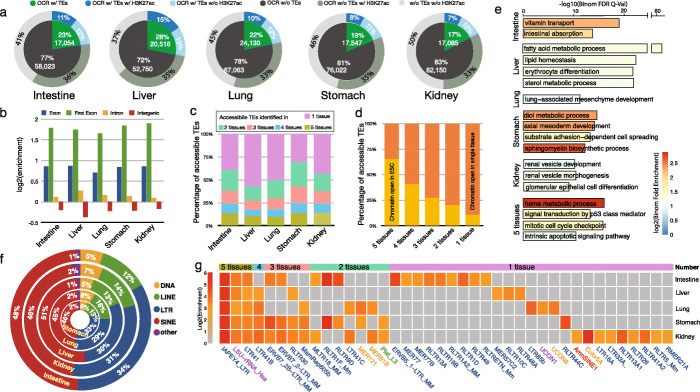
Fig. 1.
Accessible transposable elements (TEs) in five mouse tissues. a Around 17~28% of ATAC-seq peaks identified in five mouse tissues associated with TEs, near half of these accessible TEs recruited H3K27ac signals. b Genomic distribution enrichment of accessible TEs of five mouse tissues in the mouse genome. Accessible TEs highly enriched in the exons of the mouse genome, especially the first exon. c Distribution of accessible TEs among five mouse tissues. Near half of accessible TEs were identified in only one tissue type. d Chromatin status of identified accessible TEs in mouse ES cells. In total, 65% of accessible TEs constitutively in five tissues were accessible in mouse ESC, only 10% of tissue-specific TEs were open in mESC. e Gene ontology (biological process) enrichment analysis of tissue-specific accessible TEs in five mouse tissues. GO terms related to tissue-specific function were selected to visualize. f TE class distribution of accessible TEs in five mouse tissues. g TE subfamily enrichment of accessible TEs in five mouse tissues