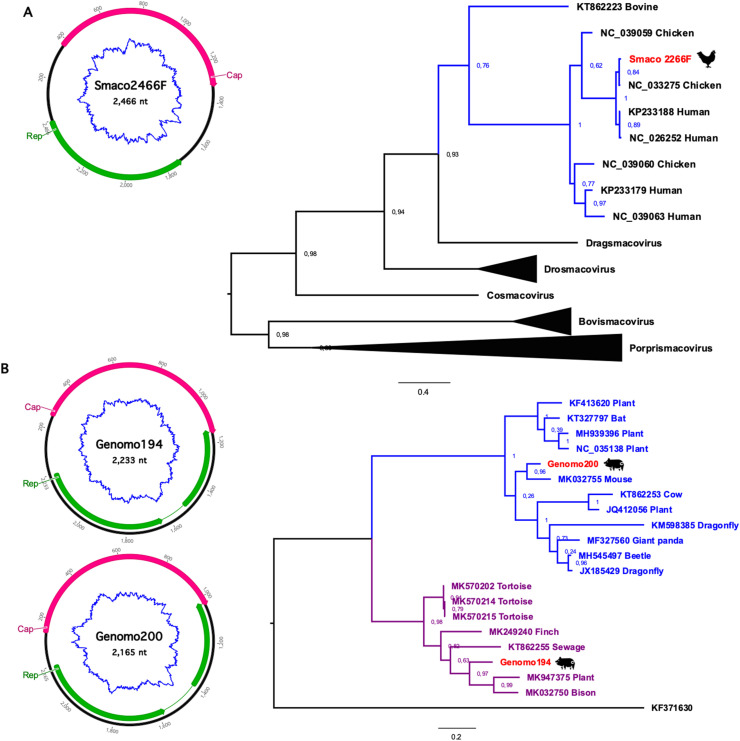
Fig. 2.
Smacoviruses and genomoviruses genomes in chicken and pork meats. (A) Genomic organization and ML phylogenetic tree of smacoviruses. Huchismacovirus genus branch are highlighted in blue. (B) Genomic architecture of genomoviruses recovered from pork samples and phylogenetic assessment. Gemycircularvirus and Gemykibivirus genus are highlighted in blue and purple color. ML phylogenetic trees were constructed using PhyML (Guindon et al., 2010) with best fit substitution models (LG_G + I + F for both sequence sets) determined by Smart Model Selection (Lefort et al., 2017). Statistical significance analyses of tree topologies were performed with the approximate likelihood branch support test (aLRT) (Guindon and Gascuel, 2003). Sequences described in this study are marked in red. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)