Abstract
Long non-coding RNAs (lncRNAs) have recently gained attention due to their important roles in human cancer types, such as breast and gastric cancer. The present study measured alterations in muskelin 1 antisense RNA (MKLN1-AS) expression in hepatocellular carcinoma (HCC) and evaluated its clinical value in patients with HCC. Additionally, the current study investigated the effects of MKLN1-AS on the malignant features of HCC cells. The detailed molecular mechanisms underlying the cancer-promoting activities of MKLN1-AS in HCC cells were also elucidated. MKLN1-AS expression in HCC tissues and cell lines was detected using reverse-transcription quantitative PCR (RT-qPCR). Cell Counting Kit-8 assays and flow cytometry were used to determine the roles of MKLN1-AS in HCC cell proliferation and apoptosis. Migration and invasion assays, as well as tumor xenograft experiments were conducted to analyze migration and invasion in vitro and tumor growth in vivo, respectively. The interaction among microRNA-654-3p (miR-654-3p), MKLN1-AS and hepatoma-derived growth factor (HDGF) in HCC was investigated using luciferase reporter assay, RNA immunoprecipitation assay, RT-qPCR, western blotting and rescue experiments. MKLN1-AS was upregulated in HCC tissues and cell lines, and a high MKLN1-AS expression was associated with shorter overall survival and disease-free survival in patients with HCC. Functionally, the knockdown of MKLN1-AS impaired HCC cell proliferation, migration and invasion, as well as induced cell apoptosis in vitro. Knockdown of MKLN1-AS expression also inhibited cell proliferation in vivo. The results indicated that MKLN1-AS functioned as a competing endogenous RNA by sponging miR-654-3p in HCC cells. Additionally, miR-654-3p targeting of HDGF was positively modulated by MKLN1-AS, and miR-654-3p knockdown partially abrogated this effect. Rescue experiments demonstrated that knockdown of miR-654-3p and overexpression of HDGF both abolished MKLN1-AS knockdown-induced cellular processes in HCC. In summary, MKLN1-AS induced pro-oncogenic effects during HCC progression by serving as a molecular sponge for miR-654-3p to increase HDGF expression. Therefore, the MKLN1-AS/miR-654-3p/HDGF axis may offer a novel target for the diagnosis, prognosis, prevention and treatment of HCC.
Keywords: apoptosis, competing endogenous RNA, muskelin 1 antisense RNA
Introduction
Hepatocellular carcinoma (HCC) is the fifth most frequent cancer type and the second most common cause of cancer-associated mortality globally (1), and HCC cases in China account for ~55% of all HCC cases worldwide (2). Every year, >780,000 novel HCC cases are reported globally, causing ~745,000 mortalities (3,4). Recent advances in the treatment of HCC, including surgical excision, liver transplantation, targeted drug therapy and chemoradiotherapy, have considerably improved the quality of life of the patient and their prognosis (5). However, the long-term clinical outcomes of patients with HCC is one of the shortest among all human cancer types (6). For instance, the survival rate of patients with HCC is <5%, with >50% cases experiencing recurrence and metastasis (7). Multiple risk factors, such as hepatitis B and C virus infections, long-term alcoholism, non-alcoholic fatty liver disease and consumption of food contaminated by Aspergillus flavus, are closely associated with HCC and its progression (8,9). However, the processes underlying HCC pathogenesis remain largely unknown and require further elucidation. Therefore, it is important to acquire an in-depth understanding of the mechanisms associated with HCC progression, which may help identify a promising strategy for tumor prevention and therapy.
Long non-coding RNAs (lncRNAs) are a diverse group of non-coding RNA transcripts comprising >200 nucleotides (10). lncRNAs do not encode proteins but are widely distributed in somatic cells, and can interact with DNA, RNA or proteins to modulate gene expression at transcriptional and post-transcriptional levels (11). A previous review has shown that lncRNAs serve important roles in numerous biological processes, including differentiation, proliferation and metabolism, and also have crucial functions in disease pathogeneses, particu-larly cancer (12). Differentially expressed lncRNAs have been observed in HCC by several studies (13–15), suggesting a crucial role of lncRNAs in HCC. Moreover, lncRNAs exhibit cancer-inhibiting or cancer-promoting activities and are implicated in the regulation of pathological processes in malignant cells (16).
MicroRNAs (miRNAs/miRs) are widely expressed in eukaryotes and are a family of endogenous, highly conserved and single-stranded small non-coding RNA molecules comprising ~23 nucleotides (17). miRNAs effectively regulate genes by directly binding to the 3′-untranslated regions (UTRs) of their target genes, resulting in mRNA degradation and/or translation inhibition (18). Previous studies have reported that numerous miRNAs contribute to HCC oncogenesis and progression (19–21). For example, miR-375 (22), miR-369-3p (23) and miR-499 (24) are weakly expressed in HCC, and execute anti-oncogenic actions. It has been revealed that lncRNAs harbor miRNA-response elements and can serve as competing endogenous RNAs (ceRNAs) or molecular sponges to modulate miRNAs and liberate their binding to target mRNAs (25).
The present study measured alterations in muskelin 1 antisense RNA (MKLN1-AS) expression in HCC and evaluated its clinical value in patients with HCC. In addition, the current study investigated the role of MKLN1-AS in regulating the malignant features of HCC cells, and the detailed molecular mechanisms responsible for the cancer-promoting actions of MKLN1-AS in HCC were elucidated.
Materials and methods
Tissue specimens
The present study was approved by the Ethics Committee of Shibo High-Tech Hospital and was performed in accordance with the Declaration of Helsinki. Written informed consent was obtained from all participants. HCC tissues and corresponding adjacent healthy tissues (≥3 cm) were collected from 65 patients (39 males; 26 females; age range, 48–72 years) admitted to Shibo High-Tech Hospital between June 2013 and February 2015. None of the patients were previously diagnosed with other human cancer types or received anticancer therapies. All tissue specimens were snap-frozen in liquid nitrogen (−196°C) immediately after tissue excision and immersed in liquid nitrogen.
Cell culture
Transformed Human Liver Epithelial-3 cells (THLE-3) and HCC cell lines (SNU-182, HuH7 and Hep3B) were acquired from the Cell Bank of the Chinese Academy of Sciences. THLE-3 cells were cultured in Bronchial Ep ithelial Growth Medium (Clonetics Corporation) that was supplemented with 10% heat-inactivated FBS (Gibco; Thermo Fisher Scientific, Inc.), 5 ng/ml epidermal growth factor and 70 ng/ml Phosphoethanolamine. The HCC cell line SNU-398 (American Type Culture Collection) was cultured in RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 ng/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc.). HuH7 and SNU-182 cells were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) and RPMI-1640, respectively, which were supplemented with 10% FBS, 1% GlutaMAX, 1% non-essential amino acids, 100 U/ml penicillin and 100 ng/ml streptomycin. Minimal essential media (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS, 1% GlutaMAX, 1% non-essential amino acids, 1% sodium pyruvate 100 mM solution, 100 U/ml penicillin and 100 ng/ml streptomycin was used to culture Hep3B cells. All cells were maintained at 37°C in a humidified atmosphere containing 5% CO2.
Cell transfection
miR-654-3p mimic and miR-654-3p inhibitor were acquired from Shanghai GenePharma Co., Ltd., and were used to upregulate and downregulate miR-654-3p expression, respectively. Negative control (NC) miRNA mimic (miR-NC) and NC inhibitor served as the control for miR-654-3p mimic and miR-654-3p inhibitor, respectively. The miR-654-3p mimics sequence was 5′-UUCCACUACCAGUCGUCUGUAU-3′ and the miR-NC sequence was 5′-UUGUACUACACAAAAGUACUG-3′. The miR-654-3p inhibitor sequence was 5′-AAGGUGAUGGUCAGCAGACAUA-3′ and the NC inhibitor sequence was 5′-ACUACUGAGUGACAGUAGA-3′. Small interfering RNAs (siRNAs) specifically targeting MKLN1-AS (si-MKLN1-AS) and NC siRNA (si-NC) were chemically synthesized by Guangzhou RiboBio Co., Ltd. The si-MKLN1-AS sequences were as follows: si-MKLN1-AS #1, 5′-TACTAAAAATACAAAAAATTAGC-3′; si-MKLN1-AS #2, 5′-AAACACTTTCAGGATATAATTGG-3′; and si-MKLN1-AS #3, 5′-GACCAAAAATGGGGATCTTTGA-3′. The si-NC sequence was 5′-CACGATAAGACAATGTATTT-3′. The hepatoma-derived growth factor (HDGF) overexpression plasmid pcDNA3.1-HDGF (pc-HDGF) and empty pcDNA3.1 plasmid were designed and generated by Shanghai GenePharma Co., Ltd.
After plating in 6-well plates with a density of 6×105 cells and incubating overnight, cells were transfected with the miRNA mimic (100 pmol), miRNA inhibitor (100 pmol), siRNA (100 pmol) or plasmid (4 μg) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) in accordance with the product specifications. Reverse-transcription quantitative PCR (RT-qPCR), flow cytometry analysis, migration and invasion assays and western blotting were conducted after 48 h incubation. A Cell Counting Kit-8 (CCK-8) assay was performed at 24 h post-transfection.
RT-qPCR
Total RNA isolation was performed using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and total RNA was quantified using a NanoDrop 1000 spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc.). For MKLN1-AS and HDGF detection, total RNA was reverse-transcribed into first-strand cDNA using PrimeScript™ RT reagent kit (Takara Biotechnology Co., Ltd.). The temperature protocol for reverse transcription was as follows: 37°C for 15 min and 85°C for 5 sec. Then, cDNA was subjected to qPCR using SYBR® Premix Ex Taq™ (Takara Biotechnology Co., Ltd.). The thermocycling conditions were as follows: Initial denaturation for 5 min at 95°C, followed by 40 cycles of 95°C for 30 sec and 65°C for 45 sec.
To determine miR-654-3p expression, cDNA synthesis and qPCR were performed using the miScript RT kit (Qiagen GmbH) and miScript SYBR Green PCR kit (Qiagen GmbH), respectively. The temperature protocols for reverse transcription were as follows: 37°C for 60 min, 95°C for 5 min and kept at 4°C. The thermocycling conditions for qPCR were as follows: Initial denaturation at 95°C for 2 min, followed by 40 cycles at 95°C for 10 sec, 55°C for 30 sec and 72°C for 30 sec. GAPDH served as the internal reference for MKLN1-AS and HDGF, and miR-654-3p expression was normalized to U6 small nuclear RNA expression. All data were analyzed using the 2-ΔΔCq method (26).
The qPCR primer sequences were as follows: MKLN1-AS forward, 5′-AAAGAGTATGTCGCTTATTGTCTAAGA-3′ and reverse, 5′-ATCCTGCTGACTTACTCCAGATGT-3′; HDGF forward, 5′-AATCAACAGCCAACAAATACCAAGT-3′ and reverse, 5′-AGCCTTGACAGTAGGGTTGTTCTC-3′; GAPDH forward, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′ and reverse, 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′; miR-654-3p forward, 5′-TCGGCAGGUGGUGGGCCGCAG-3′ and reverse, 5′-CACTCAACTGGTGTCGTGGA-3′; and U6 forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse, 5′-CGCTTCACGAATTTGCGTGTCAT-3′.
Nuclear/cytoplasmic fractionation
HCC cells (2×106 cells) were collected, and cytoplasmic and nuclear fractions were separated using a Cytoplasmic and Nuclear RNA Purification kit (Norgen Biotek Corp.). RT-qPCR was performed as aforementioned to determine relative MKLN1-AS expression in cytoplasmic and nuclear fractions.
CCK-8 assay
CCK-8 assay (Dojindo Molecular Technologies, Inc.) was performed according to the manufacturer's instructions. Transfected cells were harvested after being incubated for 24 h. A cell suspension was prepared, and its concentration was adjusted to 2×104 cells/ml. Cells were seeded into 96-well plates at a volume of 100 μl and cultured at 37°C with 5% CO2 for 0, 24, 48 and 72 h. A total of 10 μl CCK-8 reagent was added into each well, followed by an additional incubation for 2 h at 37°C with 5% CO2. The absorbance was then measured at 450 nm using a microplate reader (Bio-Rad Laboratories, Inc.).
Flow cytometry analysis
Transfected cells were digested with EDTA acid-free 0.25% trypsin and rinsed with PBS at 4°C. The cells were centrifuged at room temperature for 5 min at 12,000 × g, the supernatant was discarded and apoptosis of the collected cells was determined using an Annexin V-FITC apoptosis detection kit (Biolegend, Inc.). Cells were suspended in 100 μl binding buffer and then stained with 10 μl Annexin V-FITC and 5 μl PI at room temperature for 15 min in the dark. The percentage of early + late apoptotic cells were detected using a flow cytometer (FACScan; BD Biosciences). Data was analyzed with the CellQuest software (version 2.9; BD Biosciences).
Migration and invasion assays
For migration assays, transfected cells were collected at 48 h post-transfection and resuspended in basal medium without FBS. The upper compartments of Transwell chambers (size, 8 μM diameter; Corning, Inc.) were loaded with 200 μl cell suspension containing 5×104 cells, while 600 μl culture medium supplemented with 20% FBS was added into the lower compartments. After culturing for 24 h, the non-migrated cells remaining in the upper surface of the membrane were removed with a cotton swab, and the migrated cells were fixed with 100% methanol at room temperature for 30 min and stained with 0.5% crystal violet at room temperature for 30 min. After extensive washing with PBS, the stained cells in five random fields at ×200 magnification were imaged and counted using an inverted light microscope (Olympus Corporation). For invasion assays, 40 μl Matrigel (BD Biosciences) was used to pre-coat the Transwell chambers at 37°C for 2 h, and the aforementioned steps were repeated.
Tumor xenograft experiments
To obtain a MKLN1-AS stable knockdown HCC cell line, lentiviral expression vector stably expressing MKLN1-AS-short hairpin RNA (shRNA; sh-MKLN1-AS) and NC-shRNA (sh-NC) was acquired from Shanghai GenePharma Co., Ltd., mixed with polybrene (5 μg/ml; Sigma-Aldrich; Merck KGaA) and transfected into SNU-398 cells with MOI=5. Subsequently, 1 μg/ml puromycin was applied at 37°C to incubate the transfected cells for 4 weeks, and select MKLN1-AS stably silenced SNU-398 cells.
The experimental procedures for animal studies were approved by the Animal Experiment Administration Committee of Shibo High-Tech Hospital. Male BALB/c nude mice (n=6; age, 4–6 weeks; weight, 20 g) were obtained from Hunan SJA Laboratory Animal Co., Ltd (Hunan, China) and raised in a specific pathogen-free environment at 25°C with 50% humidity, with a 10/14-h light/dark cycle and ad libitum food and water access. A total of 5×106 SNU-398 cells stably expressing sh-MKLN1 or sh-NC were collected, resuspended in PBS and subcutaneously injected into the flanks of nude mice. The growth of tumor xenografts was monitored weekly using a caliper, and their volume was analyzed using the following formula: Volume (mm3) = 0.5 × width × length2. All nude mice were euthanized via cervical dislocation at 4 weeks post-cell injection, and tumor xenografts were resected and weighed.
Bioinformatics analysis
Gene Expression Profiling Interactive Analysis version 2.0 (GEPIA2; http://gepia.cancer-pku.cn), which included the data from The Cancer Genome Atlas (TCGA) and Genotype Tissue Expression (GTEx) projects, was used to assess the expression pattern of MKLN1-AS in HCC. The location of MKLN1-AS in cells was predicted using lncLocator (http://www.csbio.sjtu.edu.cn/bioinf/lncLocator/).
The online database StarBase version 2.0 (http://star-base.sysu.edu.cn/) was used to identify the target miRNAs of MKLN1-AS. The putative targets of miR-654-3p were predicted using miRDB (Verison 6.0; http://mirdb.org/), TargetScan (Release 7.2: March 2018; http://www.targetscan.org/vert_72) and StarBase version 2.0.
RNA immunoprecipitation (RIP) assay
RIP assays were performed using the Magna RIP™ RNA Binding Protein Immunoprecipitation kit (EMD Millipore). HCC cells (~1×107) were collected and lysed in RIP lysis buffer. After centrifugation 10,000 × g at 4°C for 5 min, cell lysates were incubated overnight at 4°C with magnetic beads conjugated to human Argonaute-2 (Ago2) or control immunoglobulin G (IgG) antibodies (1:5,000; both from cat. no. 03–110; EMD Millipore). The magnetic beads were treated with 0.5 mg/ml protease K 30 min at 55°C to digest the protein. The extracted immunoprecipitated RNA was subjected to RT-qPCR analysis for MKLN1-AS and miR-654-3p enrichment determination.
Luciferase reporter assay
The target fragments of MKLN1-AS containing the wild-type (WT) miR-654-3p binding site and mutant (MUT) MKLN1-AS fragments were constructed and subcloned into the pmirGLO luciferase reporter plasmid (Promega Corporation) to generate the recombinant luciferase reporter plasmids, MKLN1-AS-WT and MKLN1-AS-MUT. The recombinant luciferase reporter plasmids HDGF-WT and HDGF-MUT were also generated using the same experimental protocol. For reporter assays, WT or MUT luciferase reporter plasmids (0.2 μg) were transfected into HCC cells in the presence of miR-654-3p mimic (20 pmol) or miR-NC (20 pmol) using Lipofectamine® 2000. At 48 h after transfection, luciferase activity was determined using a dual-luciferase reporter assay system (Promega Corporation), and normalized to that of Renilla luciferase activity.
Western blot analysis
Transfected cells or homogenized tumor xenografts were lysed in RIPA lysis buffer (Beyotime Institute of Biotechnology), and then subjected to centrifugation 10,000 × g at 4°C for 5 min to harvest the supernatant and extract total protein. Total protein quantification was performed with a BCA Protein Assay kit (Sangon Biotech Co., Ltd.). Equal amounts of protein (30 μg) were subjected to 10% SDS-PAGE. Then, the targeted proteins were transfected to PVDF membranes. After blocking in 5% non-fat dried milk diluted in TBS-0.05% Tween-20 at room temperature for 2 h, the membranes were incubated overnight at 4°C with primary antibodies targeting HDGF (cat. no. ab128921; 1:1,000; Abcam) or GAPDH (cat. no. ab8245; 1:1,000; Abcam). The goat anti-mouse (cat. no. ab205719; 1:5,000; Abcam) and goat anti-rabbit (cat. no. ab6721; 1:5,000; Abcam) IgG horseradish peroxidase-conjugated secondary antibody was incubated with membranes at room temperature for 2 h. Subsequently, positive bands were detected using an ECL Advance western blotting detection kit (Cytiva). Quantity One software version 4.62 (Bio-Rad Laboratories, Inc.) was used for densitometry.
Statistical analysis
All data from three biological replicates for each experiment are presented as the mean ± SD. Both paired and unpaired Student's t-test was used for comparing differences between the two groups. Comparisons among multiple groups were conducted using one-way ANOVA followed by Tukey's test. Survival curves were plotted using the Kaplan-Meier method, after which the curves were compared using the log-rank test. The correlation between MKLN1-AS and miR-654-3p expression was analyzed using Pearson's correlation coefficient. P<0.05 was considered to indicate a statistically significant difference.
Results
MKLN1-AS is upregulated in HCC and associated with poor prognosis
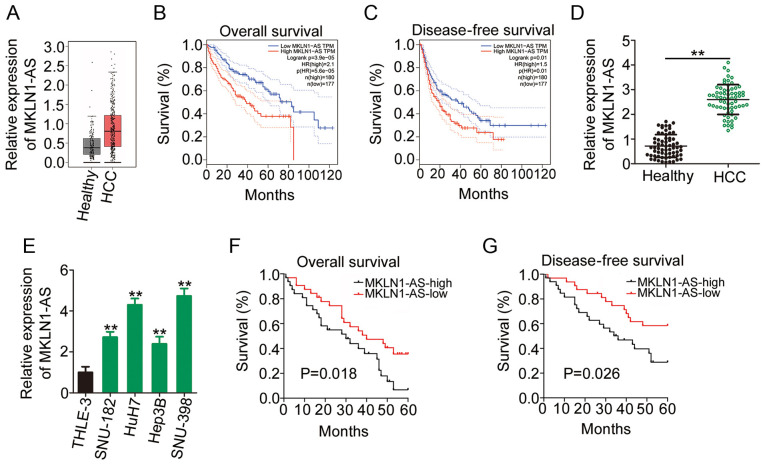
The GEPIA2 database, which includes TCGA and GTEx data, was used to assess the expression profile of MKLN1-AS in HCC. The outcomes of the box plots (Fig. 1A) demonstrated that MKLN1-AS was highly expressed in HCC tissues (n=369) compared with that in healthy liver tissues (n=160). Additionally, patients with HCC exhibiting high MKLN1-AS expression had shorter overall survival (Fig. 1B) and disease-free survival rates (Fig. 1C) compared with those exhibiting low MKLN1-AS expression, as indicated by the GEPIA2 database.
Figure 1.
MKLN1-AS is highly expressed in HCC and is associated with poor prognosis. (A) MKLN1-AS expression in HCC samples and healthy samples from TCGA and GTEx databases. TCGA and GTEx databases were used to analyze the (B) overall survival and (C) disease-free survival rates in patients with HCC exhibiting high or low MKLN1-AS expression. (D) Relative expression of MKLN1-AS in 65 pairs of HCC tissues and corresponding adjacent healthy tissues was assessed using RT-qPCR. **P<0.01 compared with adjacent healthy tissues. (E) RT-qPCR was used to measure MKLN1-AS expression in four HCC cell lines (SNU-182, HuH7, Hep3B and SNU-398) and THLE-3 cells. **P<0.01 compared with THLE-3. Kaplan-Meier analysis was used to assess the association between MKLN1-AS expression and (F) overall survival or (G) disease-free survival in the 65 patients with HCC. MKLN1-AS, muskelin 1 antisense RNA; HCC, hepatocellular carcinoma; TCGA, The Cancer Genome Atlas; RT-qPCR, reverse transcription-quantitative PCR; GTEx, Genotype Tissue Expression.
To confirm the aforementioned findings, 65 pairs of HCC tissues and corresponding adjacent healthy tissues were collected and used for MKLN1-AS quantification. RT-qPCR analysis identified that MKLN1-AS expression was higher in HCC tissues compared with adjacent healthy tissues (Fig. 1D). Similarly, MKLN1-AS expression in four HCC cell lines (SNU-182, HuH7, Hep3B and SNU-398) was increased compared with that in THLE-3 cells (Fig. 1E).
To further evaluate the clinical significance of MKLN1-AS in HCC, the median value of MKLN1-AS expression was regarded as the cutoff, and all 65 patients with HCC were classified into either MKLN1-AS-low or MKLN1-AS-high expression groups. The overall survival periods were shorter (Fig. 1F) and disease-free survival rates were lower (Fig. 1G) in the MKLN1-AS-high expression group compared with the MKLN1-AS-low expression group. Thus, these results indicated that MKLN1-AS may serve a key role in HCC oncogenesis and progression.
MKLN1-AS knockdown inhibits the proliferation, migration and invasion, as well as promotes the apoptosis of HCC cells
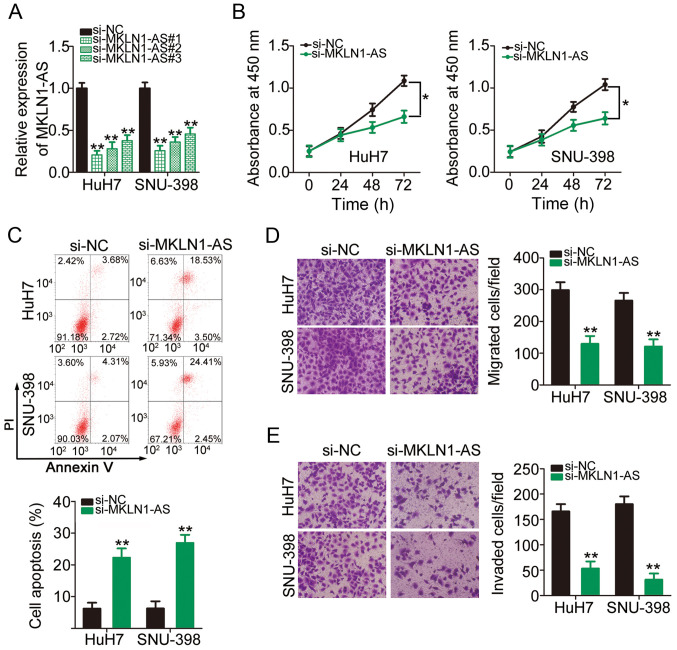
To determine the detailed role of MKLN1-AS in HCC, MKLN1-AS was knockdown in HuH7 and SNU-398 cells via transfection with si-MKLN1-AS (Fig. 2A). In particular, si-MKLN1-AS#1 exhibited the highest silencing efficacy among the three siRNAs and was therefore selected for subsequent functional assays. A CCK-8 assay was conducted to determine HCC cell proliferation. Knockdown of MKLN1-AS expression significantly suppressed HuH7 and SNU-398 cell proliferation compared with the NC (Fig. 2B). Cell apoptosis analysis demonstrated that the apoptosis of HuH7 and SNU-398 cells was significantly increased following MKLN1-AS knockdown (Fig. 2C). Moreover, the role of MKLN1-AS in the migratory and invasive abilities of HCC cells was assessed using migration and invasion assays. It was found that the migration (Fig. 2D) and invasion (Fig. 2E) of HuH7 and SNU-398 cells were inhibited after MKLN1-AS knockdown. Collectively, these results suggested that MKLN1-AS exerts tumor-promoting activities in HCC cells.
Figure 2.
Knockdown of MKLN1-AS expression inhibits hepatocellular carcinoma cell proliferation, migration and invasion, and promotes cell apoptosis in vitro. (A) Reverse transcription-quantitative PCR analysis demonstrated the efficiency of MKLN1-AS silencing in HuH7 and SNU-398 cells. (B) Proliferative ability of HuH7 and SNU-398 cells with MKLN1-AS knockdown was measured using the Cell Counting Kit-8 assay. (C) Flow cytometry analysis was used to determine the apoptotic rate of HuH7 and SNU-398 cells after MKLN1-AS knockdown. (D) Migration and (E) invasion assays were used to assess the migration and invasion of HuH7 and SNU-398 cells after MKLN1-AS silencing. *P<0.05 and **P<0.01 compared with si-NC. MKLN1-AS, muskelin 1 antisense RNA; NC, negative control; si, small interfering RNA.
MKLN1-AS acts a molecular sponge for miR-654-3p in HCC cells
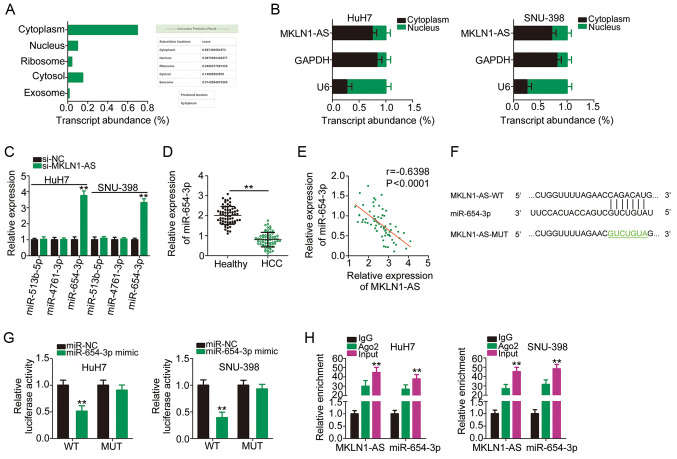
Previous studies have reported that lncRNAs execute their roles in the cytoplasm of human cancer cells by acting as small molecular sponges or ceRNAs (27,28). To determine the localization of MKLN1-AS expression, an lncRNA subcellular localization predictor, lncLocator, was used. MKLN1-AS was predicted to be mainly located in the cytoplasm (Fig. 3A), which was confirmed by the nuclear/cytoplasmic fractionation experiments (Fig. 3B). These results suggested a theoretical basis for MKLN1-AS as a sponge of miRNAs. StarBase 2.0 was used to identify the putative binding miRNAs of MKLN1-AS. It was found that MKLN1-AS has elements complementary to three miRNAs, including miR-513b-5p, miR-4761-3p and miR-654-3p. RT-qPCR analysis demonstrated that, compared with si-NC, only miR-654-3p expression was significantly increased when MKLN1-AS was knockdown (Fig. 3C). Accordingly, miR-654-3p was selected for further experimental verification.
Figure 3.
MKLN1-AS binds to and negatively modulates miR-654-3p expression in HCC cells. (A) lncLocator database predicted that MKLN1-AS was located in the cytoplasm. (B) Nuclear/cytoplasmic fractionation in combination with RT-qPCR demonstrated the distribution of MKLN1-AS in HuH7 and SNU-398 cells. (C) Expression levels of miR-513b-5p, miR-4761-3p and miR-654-3p were determined in HuH7 and SNU-398 cells after MKLN1-AS knockdown. **P<0.01 compared with si-NC. (D) miR-654-3p expression in 65 pairs of HCC tissues and corresponding adjacent healthy tissues was evaluated using RT-qPCR assay. **P<0.01 compared with adjacent healthy tissues. (E) Pearson's correlation coefficient was used to evaluate the correlation between MKLN1-AS and miR-654-3p expression levels in the 65 HCC tissues. (F) WT miR-654-3p binding site within MKLN1-AS. The sequences of MKLN1-AS containing miR-654-3p binding sequences were mutated. (G) Luciferase activity was detected in HuH7 and SNU-398 cells after co-transfection with miR-654-3p mimic or miR-NC and MKLN1-AS-WT or MKLN1-AS-MUT. **P<0.01 compared with miR-NC. (H) RNA immunoprecipitation assay was conducted to assess the binding of MKLN1-AS and miR-654-3p in HCC cells. **P<0.01 compared with IgG. WT, wild-type; MUT, mutation; MKLN1-AS, muskelin 1 antisense RNA; NC, negative control; si, small interfering RNA; miR, microRNA; Ago2, Argonaute-2; IgG, immunoglobulin G.
miR-654-3p expression was detected in the 65 pairs of HCC tissues and corresponding adjacent healthy tissues using RT-qPCR, and it was identified that miR-654-3p was downregulated in HCC tissues compared with healthy tissues (Fig. 3D). Additionally, a moderate negative correlation between miR-654-3p and MKLN1-AS expression levels was identified in the 65 HCC tissues (r=−0.6398; P<0.0001; Fig. 3E). Fig. 3F presents the predicted binding sequences of miR-654-3p within MKLN1-AS, and a luciferase reporter assay was used to validate the binding relationship between miR-654-3p and MKLN1-AS in HCC cells. The results indicated that miR-654-3p overexpression significantly decreased the luciferase activity of MKLN1-AS-WT in HuH7 and SNU-398 cells; however, a mutation at the binding site abrogated the inhibitory effect (Fig. 3G). Furthermore, compared with IgG antibody, miR-654-3p and MKLN1-AS were significantly enriched in the compound precipitated by anti-Ago2, as identified by the RIP assay (Fig. 3H). Therefore, MKLN1-AS acted as a molecular sponge for miR-654-3p in HCC cells.
HDGF is directly targeted by miR-654-3p in HCC cells and is under positive modulation of MKLN1-AS via competitive sponging of miR-654-3p
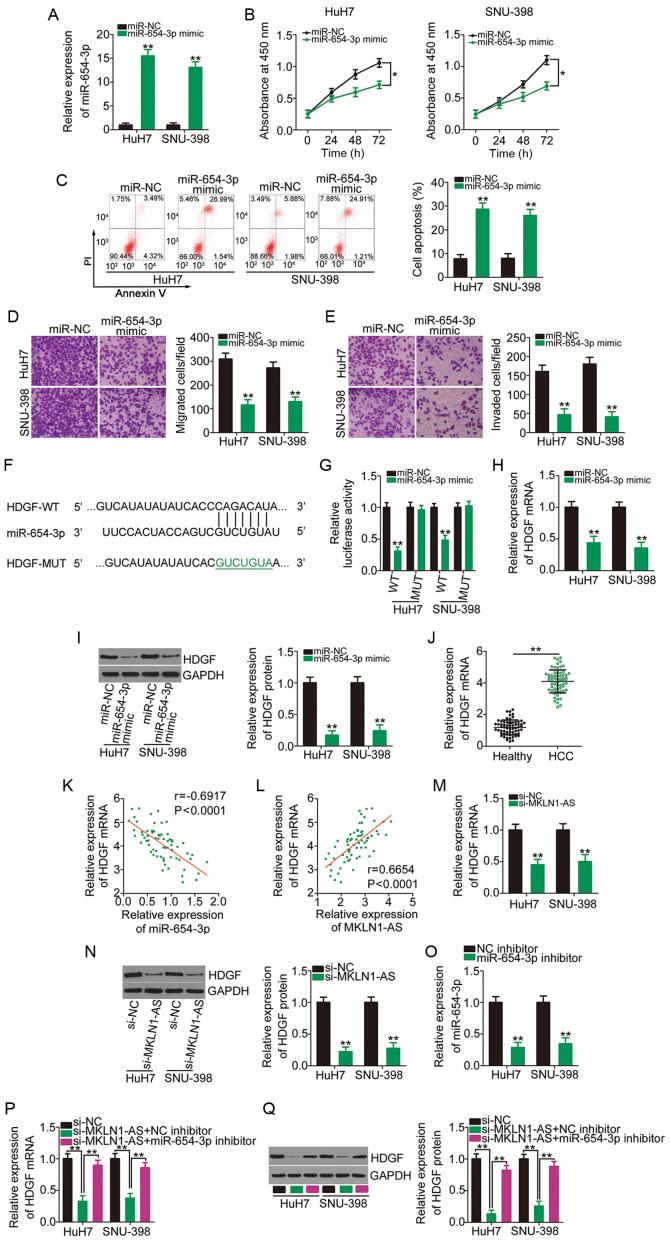
To investigate the biological roles of miR-654-3p in HCC cells, miR-654-3p was overexpressed in HuH7 and SNU-398 cells via transfection with miR-654-3p mimic (Fig. 4A). CCK-8 assay and flow cytometry analysis results demonstrated that transfection with miR-654-3p mimic in HuH7 and SNU-398 cells significantly decreased proliferation (Fig. 4B) and increased apoptosis (Fig. 4C). Using migration and invasion assays, it was observed that the number of migrated (Fig. 4D) and invaded (Fig. 4E) cells was declined in HuH7 and SNU-398 cells after miR-654-3p overexpression.
Figure 4.
MKLN1-AS positively regulates HDGF expression in HCC cells via competitive sponging of miR-654-3p. (A) HuH7 and SNU-398 cells were transfected with miR-654-3p mimic or miR-NC and subjected to RT-qPCR to determine transfection efficiency. **P<0.01 compared with miR-NC. (B) Proliferation and (C) apoptosis of miR-654-3p-overexpressing HuH7 and SNU-398 cells were evaluated using the Cell Counting Kit-8 assay and flow cytometry analysis, respectively. *P<0.05 compared with miR-NC. **P<0.01 compared with miR-NC. (D) Migration and (E) invasion assays were used to examine the migratory and invasive capacities of HuH7 and SNU-398 cells after overexpression of miR-654-3p. **P<0.01 compared with miR-NC. (F) Schematic representation of the WT and MUT complementary base pairs between miR-654-3p and the 3′-untranslated region of HDGF. (G) Luciferase activity of HDGF-WT or HDGF-MUT was determined in HuH7 and SNU-398 cells transfected with miR-654-3p mimics or miR-NC. **P<0.01 compared with miR-NC. (H) HDGF mRNA and (I) protein expression levels were quantified in HuH7 and SNU-398 cells transfected with miR-654-3p mimics or miR-NC. **P<0.01 compared with miR-NC. (J) RT-qPCR was performed to analyze HDGF mRNA expression in 65 pairs of HCC tissues and corresponding adjacent healthy tissues. **P<0.01 compared with adjacent healthy tissues. (K) Correlation between miR-654-3p and HDGF mRNA expression levels in the 65 HCC tissues was examined via Pearson's correlation coefficient. (L) Pearson's correlation coefficient analysis of MKLN1-AS and HDGF mRNA expression levels in the 65 HCC tissues. Changes in HDGF (M) mRNA and (N) protein expression levels in MKLN1-AS-depleted HuH7 and SNU-398 cells were determined using RT-qPCR and western blotting, respectively. **P<0.01 compared with si-NC. (O) Silencing efficiency of miR-654-3p inhibitor in HuH7 and SNU-398 cells was assessed using RT-qPCR. **P<0.01 compared with NC inhibitor. miR-654-3p inhibitor or NC inhibitor was transfected into HuH7 and SNU-398 cells in the presence of si-MKLN1-AS. After transfection, the (P) mRNA and (Q) protein expression levels of HDGF were measured using RT-qPCR and western blotting. **P<0.01 compared with group si-MKLN1-AS + miR-654-3p inhibitor and group si-NC. MKLN1-AS, muskelin 1 antisense RNA; NC, negative control; si, small interfering RNA; WT, wild-type; MUT, mutation; miR, microRNA; RT-qPCR, reverse transcription-quantitative PCR; HDGF, hepatoma-derived growth factor.
To elucidate the mechanism underlying the anti-oncogenic roles of miR-654-3p, the putative targets of miR-654-3p were predicted using miRDB, TargetScan and StarBase 2.0. HDGF (Fig. 4F) was identified to be a putative target of miR-654-3p, and was chosen for further study considering its oncogenic activities during HCC pathogenesis (29-31). A luciferase reporter assay was conducted to confirm the binding of miR-654-3p to the 3′-UTR of HDGF. The luciferase activity driven by HDGF-WT was significantly repressed by miR-654-3p overexpression in HuH7 and SNU-398 cells, while the luciferase activity of HDGF-MUT was unaffected in the two cell lines after co-transfection with miR-654-3p mimic (Fig. 4G). Furthermore, the results of RT-qPCR and western blotting indicated that the overexpression of miR-654-3p significantly decreased HDGF mRNA (Fig. 4H) and protein (Fig. 4I) expression in HuH7 and SNU-398 cells. It was also found that HDGF mRNA expression was significantly increased in HCC tissues (Fig. 4J) and had a moderate negative correlation with miR-654-3p expression (r=-0.6917; P<0.0001; Fig. 4K).
To investigate the ceRNA network of MKLN1-AS, the association among MKLN1-AS, miR-654-3p and HDGF was examined. First, the correlation between MKLN1-AS and HDGF mRNA expression levels was determined, and the results indicated that MKLN1-AS was moderately, positively correlated with HDGF mRNA expression in HCC tissues (Fig. 4L; r=0.6654; P<0.0001). HuH7 and SNU-398 cells were transfected with si-MKLN1-AS or si-NC, and RT-qPCR and western blotting were used to determine the changes in HDGF expression. The mRNA (Fig. 4M) and protein (Fig. 4N) expression levels of HDGF were significantly downregulated in HuH7 and SNU-398 cells after silencing MKLN1-AS.
A miR-654-3p inhibitor was used in follow-up rescue experiments, and its transfection efficiency was confirmed using RT-qPCR. Compared with the NC inhibitor, transfection with miR-654-3p inhibitor led to a significant decrease in miR-654-3p expression in HuH7 and SNU-398 cells (Fig. 4O). si-MKLN1-AS, in parallel with miR-654-3p inhibitor or NC inhibitor, was introduced into HuH7 and SNU-398 cells, and HDGF mRNA and protein expression levels in each group were determined. The mRNA (Fig. 4P) and protein (Fig. 4Q) expression levels of HDGF were significantly decreased following the transfection with si-MKLN1-AS, which was reversed by miR-654-3p inhibitor co-transfection. These data suggested that MKLN1-AS functioned as a ceRNA in HCC cells and competitively binds to miR-654-3p to increase the expression of the downstream target gene HDGF.
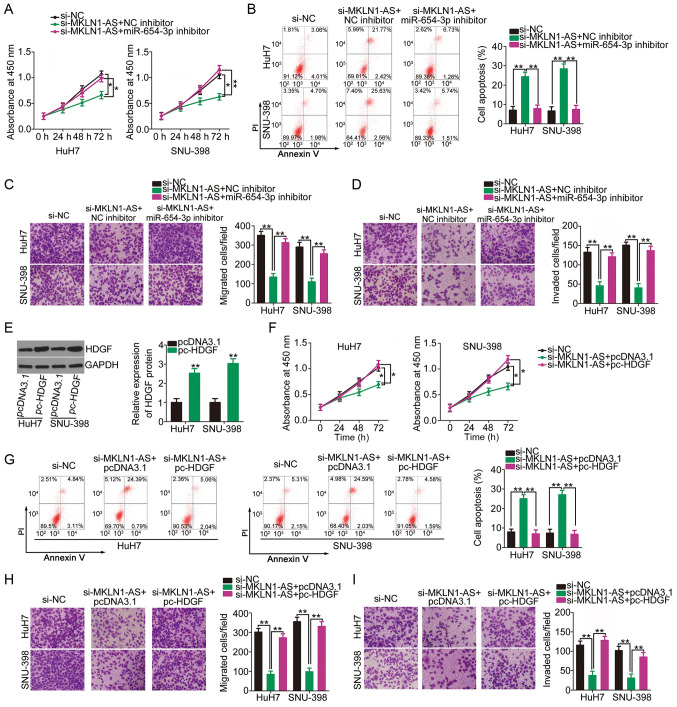
miR-654-3p inhibition and HDGF upregulation reverse the anti-oncogenic actions of MKLN1-AS knockdown in HCC cells
Rescue experiments were performed to examine whether the biological actions of MKLN1-AS in HCC cells were executed by regulating the miR-654-3p/HDGF axis. miR-654-3p inhibitor or NC inhibitor were transfected into MKLN1-AS-deficient HuH7 and SNU-398 cells. Knockdown of MKLN1-AS resulted in a significant decrease in HuH7 and SNU-398 cell proliferation (Fig. 5A) and an increase in cell apoptosis (Fig. 5B); however, these effects were alleviated by co-transfection with miR-654-3p inhibitor. Furthermore, transfection with si-MKLN1-AS significantly impaired the migration (Fig. 5C) and invasion (Fig. 5D) of HuH7 and SNU-398 cells, while these effects were reversed by the introduction of miR-654-3p inhibitor.
Figure 5.
miR-654-3p inhibition or HDGF overexpression reverses MKLN1-AS suppression-induced cellular processes in HCC cells. MKLN1-AS-deficient HuH7 and SNU-398 cells were co-transfected with miR-654-3p inhibitor or NC inhibitor. The (A) proliferation and (B) apoptosis in each group were analyzed using CCK-8 assay and flow cytometry analysis. (C) Migration and (D) invasion assays were used to determine the migration and invasion of HuH7 and SNU-398 cells that were transfected as aforementioned. (E) HuH7 and SNU-398 cells were transfected with pc-HDGF or pcDNA3.1, and the transfection efficacy was verified using western blotting. **P<0.01 compared with pcDNA3.1. (F) CCK-8 assay and (G) flow cytometry analysis were used to measure proliferation and apoptosis in HuH7 and SNU-398 cells transfected with si-MKLN1-AS along with pc-HDGF or pcDNA3.1. (H) Migration and (I) invasion of aforementioned cells were assessed using migration and invasion assays. *P<0.05 and **P<0.01. MKLN1-AS, muskelin 1 antisense RNA; NC, negative control; si, small interfering RNA; miR, microRNA; RT-qPCR, reverse transcription-quantitative PCR; HDGF, hepatoma-derived growth factor; CCK-8, Cell Counting Kit-8.
The HDGF overexpression plasmid pc-HDGF was transfected into HuH7 and SNU-398 cells, resulting in HDGF upregulation (Fig. 5E). MKLN1-AS-depleted HuH7 and SNU-398 cells were further transfected with pc-HDGF or empty pcDNA3.1 plasmid. Changes in proliferation, apoptosis, migration and invasion were then evaluated. CCK-8 assay and flow cytometry analysis demonstrated that the suppressed cell proliferation (Fig. 5F) and enhanced cell apoptosis (Fig. 5G) in cells transfected with si-MKLN1-AS were abrogated by HDGF overexpression. Additionally, the inhibitory activities of MKLN1-AS knockdown on migration (Fig. 5H) and invasion (Fig. 5I) of cells were recovered via co-transfection with pc-HDGF. Collectively, it was indicated that MKLN1-AS regulated the oncogenicity of HCC cells via the miR-654-3p/HDGF axis.
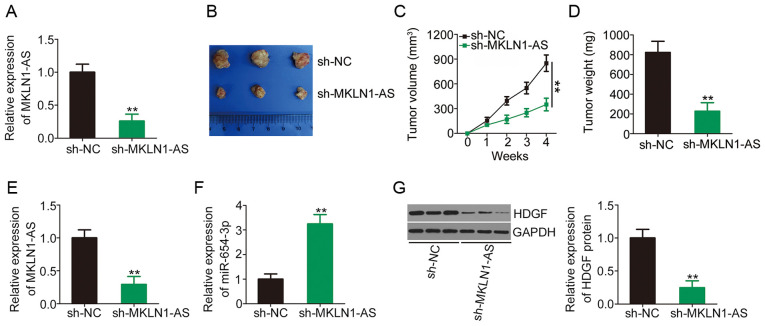
Knockdown of MKLN1-AS inhibits HCC tumor growth in vivo
To determine the functions of MKLN1-AS in HCC tumorigenesis in vivo, SNU-398 cells stably expressing sh-NC or sh-MKLN1-AS were subcutaneously inoculated into nude mice and allowed to grow for 4 weeks. The efficiency of sh-MKLN1-AS in silencing MKLN1-AS expression was determined via RT-qPCR and the results are presented in Fig. 6A. Tumor growth (Fig. 6B and C) and weight (Fig. 6D) were significantly lower in the sh-MKLN1-AS group compared with the sh-NC group. At 4 weeks post-injection, all mice were euthanized and tumor xenografts were excised to determine MKLN1-AS, miR-654-3p and HDGF expression levels. The tumor xenografts developed from MKLN1-AS depleted-SNU-398 cells had lower MKLN1-AS (Fig. 6E) and higher miR-654-3p (Fig. 6F) expression levels compared with those of the sh-NC-transfected group. Western blotting detection indicated that HDGF protein expression was significantly lower in the tumor xenografts derived from SNU-398 cells stably expressing sh-MKLN1-AS compared with the sh-NC group (Fig. 6G). These results demonstrated that MKLN1-AS silencing suppressed HCC tumor growth in vivo.
Figure 6.
Knockdown of MKLN1-AS suppresses HCC tumor growth in vivo. (A) Transfection efficiency of sh-MKLN1-AS in SNU-398 cells was analyzed via RT-qPCR. (B) SNU-398 cells infected with a lentiviral expression vector stably expressing sh-MKLN1-AS or sh-NC were subcutaneously implanted into nude mice. At 4 weeks post-injection, tumor xenografts were excised and imaged. (C) Volumes of tumor xenografts were monitored weekly, and growth curves were created. (D) Weights of tumor xenografts developed from sh-MKLN1-AS or sh-NC-transfected SNU-398 cells were detected. RT-qPCR was used to measure (E) MKLN1-AS and (F) miR-654-3p expression levels in tumor xenografts excised from sh-MKLN1-AS and sh-NC groups. (G) Expression of HDGF protein in tumor xenografts derived from SNU-398 cells stably expressing sh-MKLN1-AS or sh-NC was detected using western blotting. **P<0.01 compared with sh-NC group. MKLN1-AS, muskelin 1 antisense RNA; NC, negative control; sh, short hairpin RNA; RT-qPCR, reverse transcription-quantitative PCR; HDGF, hepatoma-derived growth factor; miR, microRNA.
Discussion
lncRNAs have received increasing attention in the past decade due to their important roles in human cancer types (32,33). Numerous lncRNAs, such as ubiquitin conjugating enzyme E2 R2-AS1 (34), colorectal neoplasia differentially expressed (35) and snail family transcriptional repressor 3-AS1 (36), are differentially expressed in HCC and are closely associated with clinicopathological factors and prognosis of patients with HCC (37–39). For instance, these lncRNAs participate in the regulation of various aggressive cellular features during hepatocarcinogenesis and cancer progression (40–42). Hence, identifying HCC-relevant lncRNAs, as well as understanding their detailed roles are critical for developing effective targets for the diagnosis, prognosis and therapy of HCC. However, to date, the functions of most cancer-associated lncRNAs in HCC remain unknown. Therefore, the aims of the present study were to determine the expression and clinical significance of MKLN1-AS in HCC, and examine the detailed roles and mechanisms of MKLN1-AS in HCC cells.
MKLN1-AS has been reported to be closely associated with the prognosis of HCC (43). In the present study, the relationship between MKLN1-AS and the clinicopathological parameters of patients with HCC was initially analyzed. It was identified that MKLN1-AS expression was upregulated in HCC, which is consistent with TCGA dataset. Analysis of TCGA data and clinical specimens also demonstrated that patients with HCC exhibiting high MKLN1-AS expression had shorter overall survival and disease-free survival rates compared with those exhibiting low MKLN1-AS expression, suggesting that high MKLN1-AS expression was associated with poor clinical outcomes. The roles of MKLN1-AS in regulating the malignant features of HCC cells in vitro and in vivo were also examined. The results suggested that MKLN1-AS exerts a carcinogenic role in HCC progression by promoting cell proliferation, migration and invasion, and inhibiting cell apoptosis in vitro. Additionally, knockdown of MKLN1-AS expression led to a decrease in HCC tumor growth in vivo. Collectively, the current results indicated that MKLN1-AS served a stimulatory role in HCC as an oncogene.
The potential mechanisms underlying the tumor-promoting activities of MKLN1-AS in HCC were investigated in the present study. It is well-established that the roles of lncRNAs depend on the cellular compartments in which they are located (44). The lncLocator database and results of nuclear/cytoplasmic fractionation experiments demonstrated that MKLN1-AS was located in the cytoplasm, which is commonly associated with ceRNA function. In general, miRNAs can be complementary to target mRNAs, leading to mRNA degradation and/or protein synthesis suppression (45). Moreover, lncRNAs can competitively bind to miRNAs and consequently modulate miRNA-mediated restriction of target mRNAs (46).
The present bioinformatics analysis identified that MKLN1-AS has complementary binding sequences for miR-654-3p. Using RT-qPCR analysis, miR-654-3p expression was found to be significantly increased after MKLN1-AS knockdown. In addition, miR-654-3p expression was downregulated in HCC tissues and negatively correlated with MKLN1-AS expression. Further study using luciferase reporter and RIP assays validated the direct binding between miR-654-3p and MKLN1-AS in HCC cells. Mechanistic investigations also demonstrated that miR-654-3p targeting of HDGF was positively modulated by MKLN1-AS, and that a miR-654-3p inhibitor partially abrogated this regulatory effect in HCC cells. Collectively, it can be concluded that MKLN1-AS increased HDGF expression in HCC cells by acting as a ceRNA and competitively binding to miR-654-3p.
miR-654-3p is downregulated in ovarian cancer (47), gastric cancer (48) and papillary thyroid cancer (49), but is upregulated in osteosarcoma (50). Moreover, miR-654-3p exerts cancer-inhibiting (47–49) or cancer-promoting roles (50), and is implicated in cancer oncogenesis and progression (47–50). A recent study reported that miR-654-3p expression was down-regulated in HCC, which was associated with poor patient prognosis, and that overexpression of miR-654-3p restricted cell proliferation, migration and invasion in HCC (51). HDGF, a heparin-binding growth factor, was first extracted from conditioned culture medium collected from the hepatoma cell line HuH7 (52) and was identified as a direct downstream target of miR-654-3p in HCC cells. HDGF performs pro-oncogenic roles in HCC and contributes to a number of malignant characteristics during HCC genesis and progression (29–31). The present results suggested that HDGF expression was directly regulated by the MKLN1-AS/miR-654-3p axis in HCC. Functional rescue experiments indicated that the miR-654-3p/HDGF axis was essential for MKLN1-AS-mediated promotion of HCC oncogenicity. Therefore, the MKLN1-AS/miR-654-3p/HDGF functional network was implicated in the development of HCC and was identified to perform important functions in various malignant processes.
In conclusion, MKLN1-AS expression is upregulated in HCC and is associated with unfavorable clinical outcomes. Additionally, it was found that knockdown of MKLN1-AS expression suppressed HCC progression. The results suggested that MKLN1-AS functioned as a ceRNA for miR-654-3p, thus increasing HDGF expression. Collectively, the present study identified a new mechanism in HCC pathogenesis and may contribute to the development of novel strategies for the diagnosis, prognosis, prevention and treatment of HCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
WJG and XHC conceived and designed the study. WJG, XHC, WC and MX carried out the experiments. WJG and XHC wrote the paper. All authors reviewed and edited the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Shibo High-Tech Hospital. The experimental procedures for animal studies were approved by the Animal Experiment Administration Committee of Shibo High-Tech Hospital. Written informed consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Goyal L, Qadan M, Zhu AX. Another treatment option for advanced hepatocellular carcinoma with portal vein thrombosis in China. JAMA Oncol. 2019;5:938–939. doi: 10.1001/jamaoncol.2019.0249. [DOI] [PubMed] [Google Scholar]
- 3.Ferlay J, Parkin DM, Steliarova-Foucher E. Estimates of cancer incidence and mortality in Europe in 2008. Eur J Cancer. 2010;46:765–781. doi: 10.1016/j.ejca.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 4.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 5.Medavaram S, Zhang Y. Emerging therapies in advanced hepatocellular carcinoma. Exp Hematol Oncol. 2018;7:17. doi: 10.1186/s40164-018-0109-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikeda K. Recent advances in medical management of hepatocellular carcinoma. Hepatol Res. 2019;49:14–32. doi: 10.1111/hepr.13259. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Q, Wang S, Qiao R, Whittaker MR, Quinn JF, Davis TP, Li H. Recent advances in magnetic nanoparticle-based molecular probes for hepatocellular carcinoma diagnosis and therapy. Curr Pharm Des. 2018;24:2432–2437. doi: 10.2174/1381612824666180516111939. [DOI] [PubMed] [Google Scholar]
- 8.Sartorius K, Makarova J, Sartorius B, An P, Winkler C, Chuturgoon A, Kramvis A. The regulatory role of microRNA in hepatitis-B virus-associated hepatocellular carcinoma (HBV-HCC) pathogenesis. Cells. 2019;8:1504. doi: 10.3390/cells8121504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zakharia K, Luther CA, Alsabbak H, Roberts LR. Hepatocellular carcinoma: Epidemiology, pathogenesis and surveillance-implications for sub-Saharan Africa. S Afr Med J. 2018;108:35–40. doi: 10.7196/SAMJ.2018.v108i8b.13499. [DOI] [PubMed] [Google Scholar]
- 10.Hu G, Niu F, Humburg BA, Liao K, Bendi S, Callen S, Fox HS, Buch S. Molecular mechanisms of long noncoding RNAs and their role in disease pathogenesis. Oncotarget. 2018;9:18648–18663. doi: 10.18632/oncotarget.24307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Egranov SD, Yang L, Lin C. Molecular mechanisms of long noncoding RNAs-mediated cancer metastasis. Genes Chromosomes Cancer. 2019;58:200–207. doi: 10.1002/gcc.22691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859:169–176. doi: 10.1016/j.bbagrm.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 13.Li W, He Y, Chen W, Man W, Fu Q, Tan H, Guo H, Zhou J, Yang P. Knockdown of LINC00467 contributed to Axitinib sensitivity in hepatocellular carcinoma through miR-509-3p/PDGFRA axis. Gene Ther. 2020 Mar 27; doi: 10.1038/s41434-020-0137-9. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 14.Lu Z, Yu Y, Ding X, Jin D, Wang G, Zhou Y, Zhu Y, Na L, He Y, Wang Q. LncRNA FLJ33360 accelerates the metastasis in hepatocellular carcinoma by targeting miRNA-140/MMP9 axis. Am J Transl Res. 2020;12:583–591. [PMC free article] [PubMed] [Google Scholar]
- 15.Liu C, Zhang M, Zhao J, Zhu X, Zhu L, Yan M, Zhang X, Zhang R. LncRNA FOXD3-AS1 mediates AKT pathway to promote growth and invasion in hepatocellular carcinoma through regulating RICTOR. Cancer Biother Radiopharm. 2020;35:292–300. doi: 10.1089/cbr.2019.3335. [DOI] [PubMed] [Google Scholar]
- 16.Chen X, Tang FR, Arfuso F, Cai WQ, Ma Z, Yang J, Sethi G. The emerging role of long non-coding RNAs in the metastasis of hepatocellular carcinoma. Biomolecules. 2019;10:66. doi: 10.3390/biom10010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu X, Tao Y, Shan L, Chen R, Jiang H, Qian Z, Cai F, Ma L, Yu Y. The role of microRNAs in hepatocellular carcinoma. J Cancer. 2018;9:3557–3569. doi: 10.7150/jca.26350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 19.Zhang T, Yang Z, Kusumanchi P, Han S, Liangpunsakul S. Critical role of microRNA-21 in the pathogenesis of liver diseases. Front Med (Lausanne) 2020;7:7. doi: 10.3389/fmed.2020.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weidle UH, Schmid D, Birzele F, Brinkmann U. MicroRNAs involved in metastasis of hepatocellular carcinoma: Target candidates, functionality and efficacy in animal models and prognostic relevance. Cancer Genomics Proteomics. 2020;17:1–21. doi: 10.21873/cgp.20163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pratama MY, Pascut D, Massi MN, Tiribelli C. The role of microRNA in the resistance to treatment of hepatocellular carcinoma. Ann Transl Med. 2019;7:577. doi: 10.21037/atm.2019.09.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li D, Wang T, Sun FF, Feng JQ, Peng JJ, Li H, Wang C, Wang D, Liu Y, Bai YD, et al. MicroRNA-375 represses tumor angiogenesis and reverses resistance to sorafenib in hepatocarcinoma. Cancer Gene Ther. 2020 Jul 3; doi: 10.1038/s41417-020-0191-x. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Q, Liu K. MiR-369-3p inhibits tumorigenesis of hepatocellular carcinoma by binding to PAX6. J Biol Regul Homeost Agents. 2020 Jun 30; doi: 10.23812/20-187-A-35. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, Li J, Chen L, Fan Z, Cheng J. MicroRNA-499 suppresses the growth of hepatocellular carcinoma by downregulating astrocyte elevated gene-1. Technol Cancer Res Treat. 2020;19:1533033820920253. doi: 10.1177/1533033820920253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song X, Cao G, Jing L, Lin S, Wang X, Zhang J, Wang M, Liu W, Lv C. Analysing the relationship between lncRNA and protein-coding gene and the role of lncRNA as ceRNA in pulmonary fibrosis. J Cell Mol Med. 2014;18:991–1003. doi: 10.1111/jcmm.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Shi SH, Jiang J, Zhang W, Sun L, Li XJ, Li C, Ge QD, Zhuang ZG. A novel lncRNA HOXC-AS3 acts as a miR-3922-5p sponge to promote breast cancer metastasis. Cancer Invest. 2020;38:1–12. doi: 10.1080/07357907.2019.1695816. [DOI] [PubMed] [Google Scholar]
- 28.Tian Y, Xia S, Ma M, Zuo Y. LINC00096 promotes the proliferation and invasion by sponging miR-383-5p and regulating RBM3 expression in triple-negative breast cancer. Onco Targets Ther. 2019;12:10569–10578. doi: 10.2147/OTT.S229659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang GY, Zhang AQ, Wang J, Li CH, Wang XQ, Pan K, Zhou C, Dong JH. Hepatoma-derived growth factor promotes growth and metastasis of hepatocellular carcinoma cells. Cell Biochem Funct. 2016;34:274–285. doi: 10.1002/cbf.3189. [DOI] [PubMed] [Google Scholar]
- 30.Enomoto H, Nakamura H, Liu W, Iwata Y, Nishikawa H, Takata R, Yoh K, Hasegawa K, Ishii A, Takashima T, et al. Down-regulation of HDGF inhibits the growth of hepatocellular carcinoma cells in vitro and in vivo. Anticancer Res. 2015;35:6475–6479. [PubMed] [Google Scholar]
- 31.Enomoto H, Nakamura H, Liu W, Nishiguchi S. Hepatoma-derived growth factor: Its possible involvement in the progression of hepatocellular carcinoma. Int J Mol Sci. 2015;16:14086–14097. doi: 10.3390/ijms160614086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agostini M, Ganini C, Candi E, Melino G. The role of noncoding RNAs in epithelial cancer. Cell Death Discov. 2020;6:13. doi: 10.1038/s41420-020-0247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsagakis I, Douka K, Birds I, Aspden JL. Long non-coding RNAs in development and disease: Conservation to mechanisms. J Pathol. 2020;250:480–495. doi: 10.1002/path.5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Z, Wei ZH, Chen SH. LncUBE2R2-AS1 acts as a microRNA sponge of miR-302b to promote HCC progression via activation EGFR-PI3K-AKT signaling pathway. Cell Cycle. 2020 Aug 23; doi: 10.1080/15384101.2020.1795991. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie SC, Zhang JQ, Jiang XL, Hua YY, Xie SW, Qin YA, Yang YJ. LncRNA CRNDE facilitates epigenetic suppression of CELF2 and LATS2 to promote proliferation, migration and chemoresistance in hepatocellular carcinoma. Cell Death Dis. 2020;11:676. doi: 10.1038/s41419-020-02853-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, Guo D, Lu G, Mohiuddin Chowdhury ATM, Zhang D, Ren M, Chen Y, Wang R, He S. LncRNA SNAI3-AS1 promotes PEG10-mediated proliferation and metastasis via decoying of miR-27a-3p and miR-34a-5p in hepatocellular carcinoma. Cell Death Dis. 2020;11:685. doi: 10.1038/s41419-020-02840-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shang R, Wang M, Dai B, Du J, Wang J, Liu Z, Qu S, Yang X, Liu J, Xia C, et al. Long noncoding RNA SLC2A1-AS1 regulates aerobic glycolysis and progression in hepatocellular carcinoma via inhibiting the STAT3/FOXM1/GLUT1 pathway. Mol Oncol. 2020;14:1381–1396. doi: 10.1002/1878-0261.12666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chi Y, Gong Z, Xin H, Wang Z, Liu Z. Long noncoding RNA lncARSR promotes nonalcoholic fatty liver disease and hepatocellular carcinoma by promoting YAP1 and activating the IRS2/AKT pathway. J Transl Med. 2020;18:126. doi: 10.1186/s12967-020-02225-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mao LH, Chen SY, Li XQ, Xu F, Lei J, Wang QL, Luo LY, Cao HY, Ge X, Ran T, et al. LncRNA-LALR1 upregulates small nucleolar RNA SNORD72 to promote growth and invasion of hepatocellular carcinoma. Aging (Albany NY) 2020;12:4527–4546. doi: 10.18632/aging.102907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin J, Xu H, Li W, Xu X, Liu H, Wei F. LINC00346 acts as a competing endogenous RNA regulating development of hepatocellular carcinoma via modulating CDK1/CCNB1 axis. Front Bioeng Biotechnol. 2020;8:54. doi: 10.3389/fbioe.2020.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen S, Wang G, Tao K, Cai K, Wu K, Ye L, Bai J, Yin Y, Wang J, Shuai X, et al. Long noncoding RNA metastasis-associated lung adenocarcinoma transcript 1 cooperates with enhancer of zeste homolog 2 to promote hepatocellular carcinoma development by modulating the microRNA-22/Snail family transcriptional repressor 1 axis. Cancer Sci. 2020;111:1582–1595. doi: 10.1111/cas.14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao JT, Chi BJ, Sun Y, Chi NN, Zhang XM, Sun JB, Chen Y, Xia Y. LINC00174 is an oncogenic lncRNA of hepatocellular carcinoma and regulates miR-320/S100A10 axis. Cell Biochem Funct. 2020 Mar 3; doi: 10.1002/cbf.3498. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 43.Xiao JR, Wang K, Liu Y, Li ZW, Zhou YJ, Wang HZ, Lu JY, Cheng SS, Wei S. Exploring of a prognostic long non-coding RNA signature of hepatocellular carcinoma by using public database. Zhonghua Liu Xing Bing Xue Za Zhi. 2019;40:805–809. doi: 10.3760/cma.j.issn.0254-6450.2019.07.014. In Chinese. [DOI] [PubMed] [Google Scholar]
- 44.Ma Y, Zhang J, Wen L, Lin A. Membrane-lipid associated lncRNA: A new regulator in cancer signaling. Cancer Lett. 2018;419:27–29. doi: 10.1016/j.canlet.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 45.Niu ZS, Wang WH, Dong XN, Tian LM. Role of long noncoding RNA-mediated competing endogenous RNA regulatory network in hepatocellular carcinoma. World J Gastroenterol. 2020;26:4240–4260. doi: 10.3748/wjg.v26.i29.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye Y, Shen A, Liu A. Long non-coding RNA H19 and cancer: A competing endogenous RNA. Bull Cancer. 2019;106:1152–1159. doi: 10.1016/j.bulcan.2019.08.011. [DOI] [PubMed] [Google Scholar]
- 47.Niu YC, Tong J, Shi XF, Zhang T. MicroRNA-654-3p enhances cisplatin sensitivity by targeting QPRT and inhibiting the PI3K/AKT signaling pathway in ovarian cancer cells. Exp Ther Med. 2020;20:1467–1479. doi: 10.3892/etm.2020.8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng G, Mou T, He J, Chen D, Lv D, Liu H, Yu J, Wang S, Li G. Circular RNA circRHOBTB3 acts as a sponge for miR-654-3p inhibiting gastric cancer growth. J Exp Clin Cancer Res. 2020;39:1. doi: 10.1186/s13046-019-1487-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geraldo MV, Nakaya HI, Kimura ET. Down-regulation of 14q32-encoded miRNAs and tumor suppressor role for miR-654-3p in papillary thyroid cancer. Oncotarget. 2017;8:9597–9607. doi: 10.18632/oncotarget.14162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou X, Li J, Teng J, Liu Y, Zhang D, Liu L, Zhang W. Long noncoding RNA BSN-AS2 induced by E2F1 promotes spinal osteosarcoma progression by targeting miR-654-3p/SYTL2 axis. Cancer Cell Int. 2020;20:133. doi: 10.1186/s12935-020-01205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang J, Chen S, Dou W, Xie R, Gao J. miR-654-3p predicts the prognosis of hepatocellular carcinoma and inhibits the proliferation, migration, and invasion of cancer cells. Cancer Biomark. 2020;28:73–79. doi: 10.3233/CBM-191084. [DOI] [PubMed] [Google Scholar]
- 52.Huang JS, Chao CC, Su TL, Yeh SH, Chen DS, Chen CT, Chen PJ, Jou YS. Diverse cellular transformation capability of overexpressed genes in human hepatocellular carcinoma. Biochem Biophys Res Commun. 2004;315:950–958. doi: 10.1016/j.bbrc.2004.01.151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.