Figure 2.
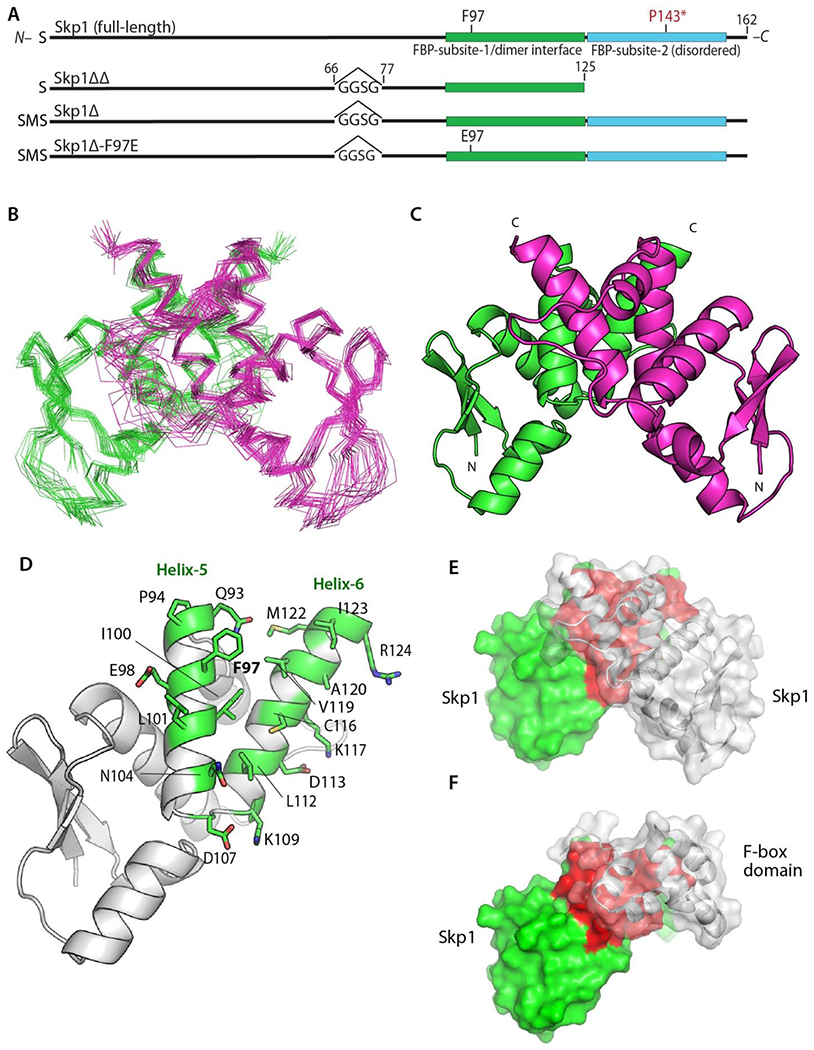
Structure of the Skp1 dimer. (A) Domain diagrams of the constructs examined. Note that versions derived from His6Skp1 have a SerMet-extension beyond the native Ser- resulting from Met removal. See Figure S1 for details. (B) Superimposition of Cα-traces of 20 calculated conformers of Skp1ΔΔ. (C) Ribbon representation of the lowest energy Skp1ΔΔ conformer (PDB ID 6V88). Dimer subunits are colored in green or magenta. A 2-fold axis of rotational symmetry lies vertically between the subunits. (D) Ribbon representation of a single Skp1ΔΔ, with the residues contributing to intermolecular contacts (<5 Å) shown in green with stick representations of their side chains. (E) Surface representation of the Skp1ΔΔ dimer is shown with the rear subunit colored in green and red, and the front in transparent gray. Red shading represents the homodimer contact region. (F) Surface representation of a hypothetical Skp1ΔΔ/F-box heterodimer model, generated by substitution of a single Skp1ΔΔ subunit for Skp1 in a human Skp1/FBXW7 complex (PDB ID 5V4B). Coloration is as in E, with FBXW7 residues 2263-2355 in gray.
